Amberlite IR-120H Catalyzed Synthesis of 1,3-Diphenylpyrazolechromenoquinolin-6-one Compounds and Their Biological Evaluation
A B S T R A C T
A series of 1,3-diphenylpyrazole-chromenoquinolin-6-one compounds were designed and synthesized by using a greener and recyclable heterogeneous Amberlite IR-120H resin as a catalyst, in the presence of ethanol reflux conditions. Interestingly, the catalyst can be recovered after completion of the reaction and can be reused without loss of catalytic property. Therefore, this method provides a green and environmentally benign much improved protocol for the synthesis of 1,3-diphenylpyrazole-chromenoquinolin-6-one compounds. The synthesized library of thirty compounds were tested against their cytotoxicity; moreover, the compounds 5s and 5t exhibited potential cytotoxic activity with IC50 values of 1.22 and 1.64 µM, respectively, on MCF-7 cancer cells. The biophysical studies such as UV-visible, fluorescence and circular dichroism studies indicate that these compounds possess good DNA intercalation ability. In addition, these compounds efficiently inhibit topoisomerase I activity. Molecular docking and viscosity studies support that these compounds exhibited intercalative mode of binding with DNA.
Keywords
Amberlite IR-120H, pyrazoles, DNA binding, anticancer, topoisomerase I, intercalation, chromenoquinoline-6-one
Introduction
Cancer is a multifaceted debilitating disease characterized by uncontrolled proliferation of cells to form tumors that represent one of the leading causes of death worldwide and has become a most common rationale in the future [1, 2]. These are different types based on cell from which organ it has originated, treatment includes chemotherapy, radiation and surgery in which chemotherapy is the mainstay. The use of existing chemotherapeutics is often narrow due to drug resistance and undesirable side effects; therefore, there is an overwhelming need to develop new agents for cancer treatment.
At present, DNA is the most essential target for the design and discovery of drugs in pharmaceutical field and many of the drug molecules currently in clinical trials are because of DNA response to their binding ability and the topological changes in DNA effective to the anticancer, anti-bacterial and therapeutic agents, the DNA binding is in different ways such as intercalation, groove binding and alkylation [3, 4]. The translation and transcription process are associated with many enzymes one such is topoisomerase I that involves in many aspects of chromatin topology and DNA metabolism. Topoisomerase inhibitors are among the noteworthy anticancer agents due to the vital role of these enzymes in cell proliferation. DNA binders like Hoechst 33258, camptothecin, netropsin and bleomycin were found to possess broad spectrum anticancer activity. Therefore, it is imperative to identify new topoisomerase inhibitors.
Heterocycles are an important and unique class of compounds. Among them, pyrazoles are best examples of aromatic heterocycles containing five membered ring incorporated with two nitrogen heteroatoms. They have attracted much attention in recent times owing to their use in agriculture and drug discovery [5]. Pyrazoles play a key role as versatile building blocks for medicinal chemistry due to their competence to exhibit broad range of bioactivities including anti-inflammatory, anti-microbial, antioxidant, anti-depressant, anti-influenza and anticancer activities [6-10].
Pyrazole derivatives are reported to exert their anticancer activity by the inhibition of multiple targets such as topoisomerase-I, -II, EGFR, fibroblast growth factor (FGF), tumor growth factor (TGF), different kinases which are significant for the management of cancer telomerase and DNA cleavage [11]. Recently, many researchers have reported numerous pyrazole derivatives, thus demonstrating the use of pyrazole scaffold in the development of new anticancer agents. Among the anticancer pyrazoles, 1,3-diphenyl pyrazoles were accounted to be efficient and highly potent cytotoxic agents as DNA binders and topoisomerase inhibitors (Figure 1) [12]. Moreover, it has been reported that pyrazole derivatives have excellent drug-like properties and high oral bioavailability [13].
Figure 1: Representative examples of DNA binders and topoisomerase inhibitors.
On the other hand, coumarins represent a key motif for the synthesis of natural products and chemotherapeutic agents. Interest in it has been amplified because, not only they are significant synthetic endpoints, but these derivatives have shown a remarkably broad spectrum of pharmacological and physiological activities and they are used as antimicrobial, antioxidant, anticancer, anti-coagulant and anti-inflammatory agents [14]. In contrast, quinoline is also an important class of compounds with great attraction for new drug design and development owing to their wide range of biological properties [15]. The polycondensed heterocyclic systems exhibited good DNA intercalation activity (Figure 1) and considering the combination principle of drug design we are interested in synthesizing the 1,3-diphenylpyrazole-chromenoquinolin-6-one compounds using a new method.
Multicomponent reactions are playing a key role in a number of organic transformations which has importance in medicinal chemistry [16]. At present multicomponent reactions are recognized as important tool for drug discovery because of their efficiency and productivity [17]. Moreover, when two different heterocyclic systems are coupled to synthesize new compounds, it is presumed that they will exhibit enhanced biological activities.
Previously the synthesis of chromenoquinolin-6-one compounds prepared via a three-component condensation of 4-hydroxy-coumarin, aldehydes and aniline was catalyzed by a sulfinic acidic ionic liquid under harsh dehydrating conditions (150oC) and a mixture of products were obtained, in other report under microwave conditions acetic acid was used as solvent [18]. However, the development of facile, greener and efficient method for the synthesis of chromenoquinolin-6ones is needed. Herein, we describe a new and efficient synthetic methodology for the preparation of 1,3-diphenylpyrazole-chromenoquinolin-6-one compounds using a solid heterogeneous acid catalyst and in vitro screening of these synthesized compounds for anticancer activity.
Results and Discussion
I Chemistry
The 1,3-diphenylpyrazole-chromenoquinolin-6-one compounds were synthesized as shown in (Scheme 1). The key aldehyde intermediates (4a-f) were prepared in two steps. Firstly, the condensation of acetophenones (1a-f) with phenyl hydrazine (2) in ethanol produced the corresponding acetophenone phenylhydrazones (3a-f). This was followed by cyclization of the acetophenone phenyl hydrazones via the Vilsmeire-Haack reaction. Finally, the condensation of subsequent aldehydes 4a-f with substituted anilines and 4-hydroxycoumarin afforded the required substituted 1,3-diphenylpyrazole-chromenoquinolin-6-one (5a-5ad) compounds in good to excellent yields.
Only a few methods are reported for the synthesis of 7,12-dihydro-6H-chromeno[4,3b] quinolin-6-one which requires high temperatures and longer reaction times. In addition, these methods produced a mixture of products, resulting in poor yields of the required product. Thus, there is still a need for the development of efficient and milder methods to overcome the shortcomings of existing protocols. In this study, we report an efficient method for the synthesis of 1,3-diphenylpyrazole-chromenoquinolin-6-one using Amberlite-IR-120H as a catalyst.
Scheme 1: Reagents and conditions: a) methanol, AcOH, rt, 5 h; b) DMF, POCl3, rt, 6 h; c) 4-hydroxy coumarin, respective aniline, Amberlite IR-120H, ethanol, reflux, 5 h.
To optimize the reaction conditions, a reaction of 4-hydroxy coumarin (1.0 mmol) with aniline (1.0 mmol) and 4a (1.0 mmol) was chosen as model reaction and the results are shown in (Table 1).
Initially, we screened different catalysts like AcOH, p-TSA, NH2SO3H, Amberlite IR120H and L-proline (Table 1, entries 1-7) to establish standard conditions. Moreover, investigation of the solvent effect on the conversion revealed that the reaction was slow in acetonitrile, chloroform, dichloromethane, water, DMF and EtOH-H2O mixture (Table 1, entries 8-13). To our delight, it was found that Amberlite IR-120H (100 mg) in ethanol refluxed for 5 h can efficiently catalyze the reaction to furnish required product (5a) with 86% yield (Table 1, entry 7). Further, the reaction was evaluated under solvent-free, catalyst-free and varying temperature conditions but without much success (Table 1, entries 14-17).
With the optimal conditions in hand, we next examined the scope of substrates on the reaction. Both electron-deficient and electron-rich substituents on aldehydes did not show much effect on the yields of the products, but the electron-deficient substituents on aniline resulted in lower yields as compared to electronrich substituents. In addition, it was observed that after completion of the reaction the catalyst was recovered, and it could be reused up to 4 cycles without loss of catalytic property. All the synthesized compounds were analyzed by 1H NMR, 13C NMR and HRMS.

|
Entry |
Catalyst |
Temperature (°C) |
Time (h) |
Solvent |
Yielda (%) |
|
1 |
p-TSA |
80 |
8 |
EtOH |
20 |
|
2 |
p-TSA |
100 |
8 |
H2O |
10 |
|
3 |
L-proline |
100 |
8 |
EtOH |
30 |
|
4 |
AcOH |
80 |
8 |
EtOH |
53 |
|
5 |
NH2SO3H |
80 |
8 |
EtOH |
65 |
|
6 |
NH2SO3H |
100 |
8 |
EtOH |
49 |
|
7 |
Amberlite IR-120 H |
80 |
5 |
EtOH |
86 |
|
8 |
Amberlite IR-120 H |
100 |
5 |
H2O |
32 |
|
9 |
Amberlite IR-120 H |
120 |
8 |
DMF |
68 |
|
10 |
Amberlite IR-120 H |
40 |
5 |
DCM |
30 |
|
11 |
Amberlite IR-120 H |
60 |
5 |
CHCl3 |
55 |
|
12 |
Amberlite IR-120 H |
80 |
5 |
ACN |
60 |
|
13 |
Amberlite IR-120 H |
85 |
8 |
EtOH-H2O |
51 |
|
14 |
Amberlite IR-120 H |
rt |
8 |
EtOH |
30 |
|
15 |
-- |
rt |
12 |
-- |
-- |
|
16 |
-- |
100 |
12 |
-- |
15 |
|
17 |
Amberlite IR-120 H |
100 |
12 |
-- |
trace |
aIsolated yields
Figure 2: Plausible mechanism for the formation of 1,3-diphenylpyrazole-chromenoquinolin-6-one compounds.
Plausible Mechanism
The plausible mechanism for the formation of 1,3-diphenylpyrazole-chromenoquinolin-6one compounds was depicted in (Figure 2). At first the pyrazole aldehyde (4) on condensation with aniline (6) resulted in the formation of imine intermediate (A), which on the nucleophilic attack of coumarin gave the intermediate B. Then, this intermediate underwent several rearrangements (C-D) and finally undergone cyclodehydration which offered the required 1,3diphenylpyrazole-chromenoquinolin-6-one (5).
II Biology
i In Vitro Anticancer Activity
All the newly synthesized compounds (5a-ad) were evaluated for their in vitro anti proliferative activity against a panel of four human cancer cell lines, such as A549 (lung), MCF7 (breast), DU-145 (prostate) and HeLa (cervical) by using MTT assay [19]. The IC50 (µM) values (concentration required to inhibit 50% of cancer cell growth) were calculated for all the tested compounds with respect to doxorubicin as positive control and the results to this regard were summarized in (Table 2). The screening results revealed that most of the compounds exhibited good to moderate cytotoxicity against the tested cancer cell lines with IC50 values ranging from 1.22 to 62.57 µM.
All the synthesized compounds showed pronounced activity against MCF-7 cell line as compared to other cell lines. Among those 5f, 5j, 5q-5t and 5x exhibited considerable cytotoxicity against breast cancer (MCF-7) cell line (<3 µM). It was noticed that the compounds 5s and 5t were found to be most effective against MCF-7 cell line among the series exhibiting potential cytotoxic activity with IC50 values of 1.22 and 1.64 µM, respectively, and have also shown considerable cytotoxicity against the other tested cancer cell lines.
Table 2: Cytotoxicity of 1,3-diphenylpyrazole-chromenoquinolin-6-one compounds (5a‒ad) on selected human cancer cell lines.
|
|
Compound |
|
|
|
|
|
IC50 values (µM) a |
|
|
|
|
||||
|
|
|
|
A549b |
|
|
MCF-7c |
|
|
HeLad |
|
|
DU-145e |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
5a |
21.26 ± 1.08 |
|
9.91 ± 0.55 |
|
22.38 ± 1.55 |
|
18.32 ± 0.88 |
|
|
|||||
|
|
5b |
26.91 ± 1.92 |
|
8.20 ± 0.42 |
|
22.96 ± 1.32 |
|
17.51 ± 0.92 |
|
|
|||||
|
|
5c |
32.35 ± 1.86 |
|
10.73± 0.34 |
|
39.90 ± 2.22 |
|
21.85 ± 0.86 |
|
|
|||||
|
|
5d |
50.11 ± 2.78 |
|
15.27 ± 0.46 |
|
62.57 ± 3.96 |
|
30.85 ± 1.78 |
|
|
|||||
|
|
5e |
6.46 ± 0.99 |
|
5.46 ± 0.51 |
|
14.79 ± 2.02 |
|
6.31 ± 0.99 |
|
|
|||||
|
|
5f |
10.81 ± 0.63 |
|
1.98 ± 0.71 |
|
3.23 ± 0.22 |
|
4.69 ± 0.63 |
|
|
|||||
|
|
5g |
11.32 ± 0.89 |
|
3.68 ± 0.91 |
|
14.12 ± 1.02 |
|
6.53 ± 0.89 |
|
|
|||||
|
|
5h |
25.50 ± 1.54 |
|
11.70 ± 0.61 |
|
25.11 ± 1.86 |
|
18.01 ± 1.54 |
|
|
|||||
|
|
5i |
19.66 ± 0.98 |
|
5.02 ± 0.71 |
|
18.38 ± 1.04 |
|
10.78 ± 0.98 |
|
|
|||||
|
|
5j |
15.55 ± 1.52 |
|
1.82 ± 0.24 |
|
4.68 ± 0.31 |
|
6.84 ± 0.52 |
|
|
|||||
|
|
5k |
15.24 ± 1.26 |
|
4.68 ± 0.46 |
|
18.72 ± 1.02 |
|
9.14± 1.26 |
|
|
|||||
|
|
5l |
16.98 ± 0.52 |
|
3.65 ± 0.23 |
|
15.70 ± 1.03 |
|
6.61 ± 0.52 |
|
|
|||||
|
|
5m |
16.73 ± 1.06 |
|
3.50 ± 0.96 |
|
7.24 ± 0.98 |
|
10.47 ± 1.86 |
|
|
|||||
|
|
5n |
39.81 ± 2.05 |
|
12.56 ± 1.66 |
|
19.67 ±1.96 |
|
21.92 ± 1.26 |
|
|
|||||
|
|
5o |
24.83 ± 1.52 |
|
17.17 ± 1.03 |
|
19.61 ± 2.02 |
|
20.18 ± 0.64 |
|
|
|||||
|
|
5p |
14.21 ± 1.29 |
|
11.81 ± 0.94 |
|
21.72 ± 1.03 |
|
11.22 ± 1.69 |
|
|
|||||
|
|
5q |
13.30 ± 1.38 |
|
2.75 ± 0.54 |
|
4.26 ± 0.76 |
|
6.18 ± 0.96 |
|
|
|||||
|
|
5r |
15.88 ± 1.62 |
|
2.80 ± 0.84 |
|
4.36 ± 0.12 |
|
6.92 ± 0.47 |
|
|
|||||
|
|
5s |
10.59 ± 0.43 |
|
1.22 ± 0.94 |
|
3.51 ± 0.13 |
|
2.99 ± 0.13 |
|
|
|||||
|
|
5t |
5.25 ± 0.23 |
|
1.64 ± 0.76 |
|
2.71 ± 0.33 |
|
4.52 ± 0.37 |
|
|
|||||
|
|
5u |
19.05 ± 2.69 |
|
11.41 ± 0.52 |
|
17.37 ± 0.02 |
|
13.58 ± 2.69 |
|
|
|||||
|
|
5v |
17.11 ± 1.36 |
|
3.23 ± 0.98 |
|
10.56 ± 0.41 |
|
7.49 ± 0.76 |
|
|
|||||
|
|
5w |
21.00 ± 1.65 |
|
9.75 ± 0.33 |
|
16.18 ± 0.66 |
|
15.89 ± 1.65 |
|
|
|||||
|
|
5x |
15.88 ± 1.20 |
|
2.81 ± 0.87 |
|
4.36 ± 0.53 |
|
6.92 ± 0.60 |
|
|
|||||
|
|
5y |
11.35 ± 1.23 |
|
9.53 ± 0.91 |
|
23.77± 1.89 |
|
10.30 ± 1.23 |
|
|
|||||
|
|
5z |
26.30 ± 2.36 |
|
13.92 ± 0.78 |
|
24.85 ± 2.49 |
|
19.56 ± 1.36 |
|
|
|||||
|
|
5aa |
28.84 ± 2.13 |
|
20.14 ± 1.02 |
|
26.91 ± 1.26 |
|
22.40 ± 1.73 |
|
|
|||||
|
|
5ab |
17.37 ± 0.95 |
|
12.73 ± 0.51 |
|
16.87 ± 1.23 |
|
14.12 ± 0.61 |
|
|
|||||
|
|
5ac |
17.78 ± 1.05 |
|
13.66 ± 1.56 |
|
21.18 ± 2.01 |
|
17.00 ± 1.05 |
|
|
|||||
|
|
5ad |
20.41 ± 0.92 |
|
12.30 ± 1.32 |
|
20.46 ± 1.36 |
|
18.31 ± 0.76 |
|
|
|||||
|
|
Doxorubicin |
1.24 ± 0.59 |
|
1.32 ± 0.11 |
|
1.53 ± 0.47 |
|
2.11 ± 0.70 |
|
|
|||||
|
|
(Positive control) |
|
|
|
|
|
|||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
a 50% Inhibitory concentration after 48 h of drug treatment and the values are average of three individual experiments; bHuman lung cancer; cHuman breast cancer; dHuman cervical cancer; dHuman prostate cancer.
Figure 3: Structure activity relationship of 1,3-diphenylpyrazole-chromenoquinolin-6-one compounds.
In order to know the structure-activity relationship (SAR) of the compounds, we have varied the different substitution pattern of both anilines and pyrazole aldehydes of these compounds. The electron deficient substituents like Cl and NO2 (5s and 5t) on pyrazole ring increased the activity when compared with electron rich substitution, OMe (5p) group. In contrast, the electron-rich substituents on aniline increased the activity as compared to the electron-deficient substituents (Figure 3). Among them, the 3,5-disubstituted compounds like 5s and 5t displayed promising cytotoxic activity, particularly against MCF-7 cell line.
It was observed that mono, 3,4-disubstituterd as well as trisubstituted compounds exhibited comparatively less activity then the 3,5-disubstituted compounds. These SAR studies also suggest that electron-deficient substituents like Cl and NO2 on aryl ring of the 1,3-disubstituted pyrazole subunit (R1) and electron-rich substituents like 3,5-dimethoxy on aniline (R2) were required for the cytotoxic potential. Based on the MTT assay results the best two leads among the series were further investigated for their DNA binding ability, topoisomerase I inhibition and molecular docking studies.
ii DNA Binding Studies
In addition, the exploration of biological activities of these active compounds (5s and 5t) and to understand the nature of interaction with DNA, spectroscopic studies were performed.
a UV-Visible Studies
In order to investigate the nature of binding of these compounds to DNA, the UV absorption spectra of active synthesized compounds in the presence and absence of increasing concentrations of calf thymus (CT) DNA were examined (Figure 4). On addition of DNA to compounds solution, the absorption band at 357 nm and 332 nm intensity gradually reduced without exhibiting any shift. Since the absorption bands shows hypochromicity upon addition of DNA, this indicate that the 5s and 5t compounds interact well with DNA. UV-visible titration studies alone were not sufficient to understand the interaction between synthesized compounds and CT DNA.
Figure 4: UV-visible spectra depicts the interaction of synthesized compounds at 25 oC a) 5s (1 ml of 25.0 μM) and b) 5t (1 ml of 25.0 μM) with the addition of 5 μL of DNA (25.0 μM) each time.
b Fluorescence Spectroscopy
Fluorescence titration is another valuable technique to understand the binding mode of small molecules to DNA at comparatively lower concentration [20]. As these compounds showed fluorescence properties, their interaction with CT-DNA could provide certain observations regarding the nature of their interactions with DNA. In this study, when CT-DNA was added to the compounds, the fluorescence emission peak of 5s and 5t compounds exhibited hyperchromicity. Hyperchromicity of the fluorescence emission peak was observed at 282 nm (5s), 291 nm (5t) compounds, respectively, indicating that the fluorescence of each derivative was getting enhanced probably due to its intercalation between the two stacks of CT-DNA. In the emission spectra of the compounds, with increasing CT-DNA concentration, the emission intensity increased due to the exchange of fluorescence energy between the bases and the compound [21]. Fluorescence titration results indicated that these compounds may be intercalating between the bases of DNA. The fluorescence intensity increases when the compounds 5s and 5t come close to the DNA bases upon intercalation. The fluorescence spectra recorded on the interaction of 5s and 5t with CT-DNA is depicted in (Figure 5).
Figure 5: Fluorescence spectra indicates the interaction of compounds at 25 oC a) 5s (10 µM) and b) 5t (10 µM) with increasing concentrations of CT-DNA (multiples of 0.5 µM).
c Circular Dichroism Spectroscopy
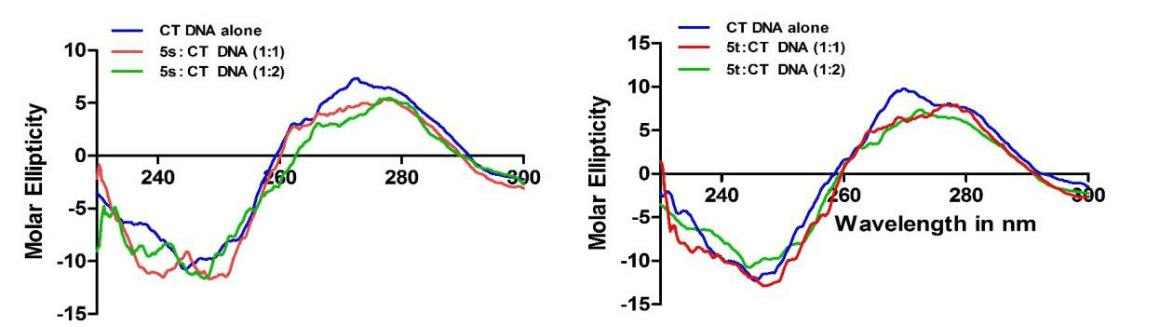
The circular dichroism (CD) is a potential technique to study the nucleic acid conformational change due to the interaction of DNA with molecules or changes in the environmental conditions. The CD spectrum of the CT DNA exhibited a positive band at 275 nm and a negative band at 245 nm due to π–π base stacking and right-hand helicity. On addition of 5s and 5t at a concentration of 10 µM to the same concentration of CT DNA solution (DNA: 5s and 5t, 1:1), positive band at 275 nm exhibited slight hypochromicity, which is an indication of melting of the DNA-compound complex on compound intercalation with DNA [22]. On further increasing the concentration of 5s and 5t compounds (DNA: 5s and 5t, 1:2) the positive band at 275 nm decreased further in its intensity, indicating further unwinding of dsDNA by compound through intercalation. The negative band intensity at 245 nm altered by the addition of 5s and 5t compounds indicated the ability of the compound to bring about change in the DNA helix. A representative figure is shown in (Figure 6).
Figure 6: Circular dichroism studies illustrate the changes in the CT DNA conformation upon interaction with complexes 5s/5t. A) CD spectra obtained with the CT DNA in 100 mM TE (pH 7.0) at 20 °C and upon addition of 1:1 and 1:2 CT DNA: complex 5s ratios. B) CD spectra obtained with the CT DNA in 100 mM TE (pH 7.0) at 20 °C and upon addition of 1:1 and 1:2 CT DNA: complex 5t ratios. In both titration reactions to 10 μM CT DNA, 10 μM and 20 μM of complexes 5s/5t was added (1:1 and 1:2 CT DNA: complexes 5s/5t ratios) and CD spectra were recorded. CD spectra were averaged over three scans. The arrows indicate the direction of movement of CD peaks upon addition of complexes 5s/5t.
From the DNA binding data, it is evident that these compounds were interacting with DNA and they may be binding to DNA and the 5s and 5t compounds may be coming close to the DNA bases.
d Viscosity Studies
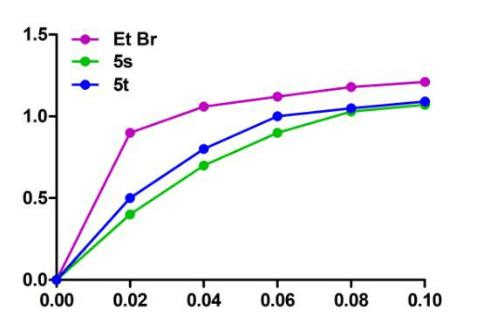
In order to clearly understand the nature of interaction of these potential molecules with CT DNA, viscosity studies were carried out. Spectroscopic studies indicated that these compounds interacted well with CT DNA. Relative viscosity of DNA increases when a molecule intercalates with DNA. The enhancement of viscosity is due to increase in the axial length of DNA helix on molecule’s intercalation. Hence, intercalation of a compound results in increase of the viscosity of DNA solutions [23]. But, reduction in the relative viscosity is typically observed with covalent DNA binding [24]. It is reported that viscosity changes will be less or will not change when molecules binds to the surface of DNA [25].
Figure 7: Effect of 5S and 5t complexes (15 µM) on the DNA viscosity was studied. Viscosity of EtBr- CT DNA (150 µM) was considered as control.
When complex intercalates with DNA, the axial length of DNA helix will increase to accommodate the binding derivatives, which leads to the increase of DNA solution viscosity. With the addition of synthesized compounds, moderate enhancement in the viscosity of DNA solution was noticed. As shown in (Figure 7), on interaction of 5s and 5t compounds with CT DNA exhibited an enhancement in the viscosity which was not as pronounced as observed for classical intercalator like ethidium bromide. Ethidium bromide (EtBr) used as control was added to CT-DNA and the viscosity of DNA was measured under similar conditions. The graphs obtained with 5s and 5t compounds were observed close to ethidium bromide (a well-known intercalator). The results obtained from viscosity experiments indicated that these compounds may show intercalative mode of interaction with DNA. The graph plotted between (ŋ/ŋo)1/3 and complex/CT-DNA is shown in (Figure 7).
iii Topoisomerase Inhibition Assay
The Topoisomerase I (topo I) binds to double stranded supercoiled DNA and cuts the single stranded portion. This will release the superhelical tension in DNA transforming it into relaxed form. Recently, DNA topo I inhibitors have gained much importance in cancer chemotherapy since the small molecules which inhibit DNA topo I activity will cause single and double stranded breaks in DNA that leads to damage of genome integrity. Since the abundance of topo I is usually high in cancer cells as compared to normal cells, DNA topo I has been identified as the potential target for several anticancer drugs used in clinical studies. Topo-I inhibitors stabilized and accumulated the DNA-topoisomerase cleavage complexes and inhibited the DNA relegation step, thus resulting in cleavage of DNA strand [26].
In the present study, the effect of compounds 5s and 5t towards inhibition of DNA topo I was studied. As discussed above, the 5s and 5t showed good interaction with DNA, by binding to DNA topoisomerase I, cleaved the complex and interfered with its religation step. Cao and coworkers have recently reported that camptothecin inhibited DNA topo I at 100 µM concentration whereas simple β-carboline harmine derivatives inhibited DNA topo I at 150 µM [27].
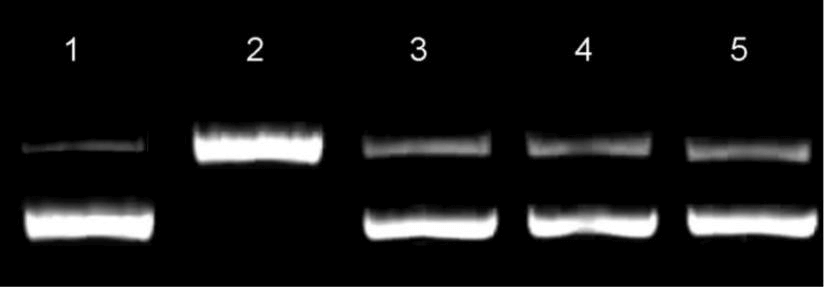
Figure 8: Effect of 20 µM of compounds 5s and 5t on transforming the supercoiled DNA (0.5 µg of pBR322 plasmid DNA) was studied in the presence and absence of 5s and 5t complexes. Camptothecin was considered as control. Lane 1: pBR322 plasmid DNA alone, lane 2: Topo I, lane 3: 5s, lane 4: 5t and lane 5: Camptothecin were loaded in each lane.
Generally, the molecules with low IC50 values including better intercalation with DNA usually exhibited superior DNA topo I inhibition. Interestingly, in the present study 5s and 5t showed lower IC50 values and considerably higher interaction with DNA. Hence, these compounds were studied for DNA topo I inhibition assay using pBR322 plasmid DNA. The compounds 5s and 5t showed considerably good DNA topoisomerase I inhibition at 10 µM concentration (Figure 8). While, the topo I has completely converted the supercoiled pBR322 plasmid DNA to linear form in the absence of 5s and 5t (lane B, Figure 8). Further comparison of the results obtained will illustrate that both 5s and 5t showed almost similar inhibition when compared with a known topo I inhibitor, namely Camptothecin (lane E). Based on these characteristic properties such as DNA intercalation and DNA topo I inhibition, the compounds 5s and 5t could be considered as potential candidates for anticancer therapy.
III Molecular Docking
Docking studies were performed to obtain an insight into the mode of binding of the active compounds (5s and 5t) to the protein and the DNA ternary complex. The coordinates of the protein were obtained from the Protein Data Bank (PDB ID: 1SC7) and necessary corrections to the protein structure were carried out using the Protein Preparation Wizard in Schrodinger Suite 2014-3. 3D structures were generated by Schrodinger suite and Schrodinger’s LigPrep program was used to generate different conformations of ligands. Molecular docking studies were performed by using a GLIDE docking module of Schrodinger suite and the results were analyzed on the basis of GLIDE docking score and molecular recognition interactions. All the 3D figures were obtained using Schrodinger Suite 2014-3 [28]. Figure 9 illustrated that these compounds fit well into M38 binding pocket present in DNA-Topo 1 complex (PDB ID: 1SC7) [29].
The docking pose for 5s showed that there is a hydrogen-bonding interaction between the carbonyl of a chromenoquinolin-6-one ring and the guanidine group of Arg364 similar to the co-crystal ligand; besides this, it also showed π-π interactions with the dC112. The N-phenyl pyrazole substitution showed pi-cation interaction with the Lys352. Whereas the chromone part of 5t showed a hydrogen-bonding interaction with DNA base pair dC112. In addition to this, it also possesses an additional hydrogen bonding interaction between the nitro group of the 4-phenyl pyrazole ring and the amino group of the side chain of Asn722. The nitro group of 5t showed π-cation interaction with the nitrogen bases dT10; on the other hand, pyrazole substitution showed π-π interactions with dT10 and dC112 nitrogen bases.
Furthermore, we observed hydrophobic interactions with amino acids close to the binding site, in which the pyrazole substitution of 5s showed interaction with the Asp533, Ile535, Leu721, Asn722 and the chromenoquinolin-6-one ring with Gln365, Ala351, Asn352, Lys425. Compound 5t showed some of the key hydrophobic interactions with amino acid residues Arg364, Gln356, Lys374, Lys532, Thr718, Asn722, Ala351, Asn352, Lys425, Tyr426, Ile427, Met428 and Lys436, which stabilizes the compound 5t in the active pocket. These studies indicated that the 1,3-diphenylpyrazole-chromenoquinolin-6-one compounds were capable of fitting properly in the binding site of the DNA topoisomerase I.
The biophysical studies already showed that these compounds possess DNA intercalation properties. Therefore, docking studies were performed to obtain a better insight into the binding mode of these compounds to the DNA. The coordinates of the DNA were obtained from the Protein Data Bank (PDB ID 1NAB) [30]. Docking studies showed that 5s and 5t bind to the DNA through intercalation. The docking pose of 5s and 5t showed that the chromenoquinolin-6-one ring lies in the central part of the DNA intercalation cavity and was stacked between the dC1, dG2 of chain A and dG12 of chain B, while N-phenyl pyrazole substitution of 5s interacted with dA-3 and 5t interacted with dG12 nucleotides present in the groove (Figure 10). The N-phenyl substitution extended towards the minor groove and the chromenoquinolin-6-one substitution towards the major groove. Docking studies on DNA topoisomerase-I and CT DNA showed that these compounds have the potential to bind to the DNA Topo-I and also intercalated with DNA.
Figure 9: Docking poses for 5s and 5t in DNA Topo I (PDB ID: 1SC7); hydrogen bonds are shown in yellow dotted lines, and hydrogen bonding residues are shown in yellow and faded green colour and nucleic acid residues which are showing pi-cation interaction are shown in faded teal colour.
Figure 10: Docking poses for 5s and 5t in DNA showing the intercalation-binding mode.
Conclusion
In conclusion, we have developed an operationally simple and eco-friendly method for the synthesis of functionalized 1,3-diphenylpyrazole-chromenoquinolin-6-one compounds using the solid phase resin catalyzed condensation of 4-hydroxycoumarin, aniline and 1,3diphenylpyrazole aldehydes. The synthesized series of compounds were evaluated for their cytotoxicity, among them 5s and 5t displayed promising cytotoxic activity with IC50 values of 1.22 and 1.64 µM against human breast cancer cell line (MCF-7). Further, these compounds inhibited DNA topoisomerase I activity and spectroscopic studies indicated that these compounds intercalate with DNA. Additionally, the molecular docking and viscosity studies supported the intercalation mode of binding of these compounds with DNA. Thus, these compounds could be considered as DNA and topoisomerase I targeted potential scaffolds in favor of the development of newer leads for the treatment of breast cancer.
Experimental Section
I Materials and Methods
Chemistry: All chemicals and reagents were purchased from commercial source and were used as such without purification. Reactions were monitored by TLC on silica gel glass plate containing 60 GF-254 and visualization was achieved by UV light or iodine indicator. 1H and 13C NMR spectra were determined in CDCl3 + DMSO-d6 by using Varian and Avance instruments. Chemical shifts were expressed in parts per million (δ in ppm) downfield from TMS expressed as internal standard and coupling constants are expressed in Hz. 1H NMR spectral data were reported in the following order: multiplicity (s, singlet; brs, broad singlet; d, doublet; dd, doublet of doublets; t, triplet; m, multiplet), coupling constants in Hz and number of protons. ESI mass spectra were recorded on a Micromass Quattro LC using ESI+ software with capillary voltage 3.98 kV and an ESI mode positive ion trap detector. Melting points were determined with an electrothermal melting point apparatus and were uncorrected.
II General procedure for the synthesis of 1,3-diphenyl-1H-pyrazole-4-carbaldehydes (4ae)
To a stirred solution of substituted acetophenone (1a-e, 2 mmol) in anhydrous methanol, phenylhydrazine (2, 2.5 mmol) and acetic acid (3-4 drops) were added and allowed the reaction to stir at room temperature for the formation of acetophenone phenylhydrazones. After the completion of the reaction, the reaction mixture was evaporated under vacuum and the crude product was directly used for next step, the acetophenone phenylhydrazones (3a-e) were dissolved in DMF (5 mL), was added POCl3 (1 mL) slowly by maintaining 0 °C, and allowed the reaction at room temperature for 6 h. After completion of the reaction neutralized the reaction mixture with Na2CO3 solution, filtered the resulting crude solid and purified by column chromatography to obtain the pure intermediate aldehydes (4a-e) in good yields.
i 3-(4-methoxyphenyl)-1-phenyl-1H-pyrazole-4-carbaldehyde (4a)
White solid of 89 % yield; 1H NMR (400 MHz, CDCl3) δ 10.03 (s, 1H), 8.51 (s, 1H), 7.80 – 7.77 (m, 4H), 7.51 – 7.47 (m, 2H), 7.39 – 7.35 (m, 1H), 7.04 – 7.00 (m, 2H), 3.86 (s, 3H); MS (ESI): m/z 279 [M + H]+.
ii 3-(4-nitrophenyl)-1-phenyl-1H-pyrazole-4-carbaldehyde (4b)
Yellow solid of 85 % yield; 1H NMR (500 MHz, CDCl3) δ 10.06 (s, 1H), 8.55 (s, 1H), 7.88 – 7.82 (m, 2H), 7.70 – 7.76 (m, 2H), 7.58 – 7.50 (m, 4H), 7.46 – 7.43 (m, 1H); MS (ESI): m/z 294 [M + H]+.
iii 1,3-diphenyl-1H-pyrazole-4-carbaldehyde (4c)
White solid of 91 % yield; 1H NMR (400 MHz, CDCl3) δ 10.06 (s, 1H), 8.55 (s, 1H), 7.84 – 7.78 (m, 4H), 7.53 – 7.47 (m, 5H), 7.39 (t, J = 7.4 Hz, 1H); MS (ESI): m/z 249 [M + H]+.
iv 3-(4-chlorophenyl)-1-phenyl-1H-pyrazole-4-carbaldehyde (4d)
White solid of 82 % yield; 1H NMR (500 MHz, CDCl3) δ 10.05 (s, 1H), 8.55 (s, 1H), 7.86 – 7.84 (m, 2H), 7.81 – 7.79 (m, 2H), 7.55 – 7.48 (m, 4H), 7.44 – 7.41 (m, 1H); MS (ESI): m/z 283 [M + H]+.
v 3-(3,4-dichlorophenyl)-1-phenyl-1H-pyrazole-4-carbaldehyde (4e)
White solid of 79 % yield; 1H NMR (400 MHz, CDCl3) δ 10.04 (s, 1H), 8.53 (s, 1H), 8.07 (d, J = 2.0 Hz, 1H), 7.80 – 7.77 (m, 3H), 7.53 (dd, J = 8.4, 6.5, 2.8 Hz, 3H), 7.44 – 7.40 (m, 1H); MS (ESI): m/z 317 [M + H]+.
III General Procedure for the Synthesis of Compounds (5a-5ad)
To a stirred solution of 1,3-diphenylpyrazole aldehyde (1 mmol) in ethanol, aniline (1 mmol) and Amberlite IR-120H (100 mg) were added and continued to stir for 30 min at 80 °C, then was added coumarin. Refluxed the reaction mixture for 5 h, the completion of the reaction was monitored by TLC analysis and then filtered the reaction mixture. To the resulting crude solid ethyl acetate and water were added, the organic layer was separated, dried over anhydrous Na2SO4 and concentrated under vacuum. Finally, the compound was purified by column chromatography to afford the pure product in good yields. The catalyst in aqueous layer was collected, washed with water 2-3 times and dried in oven to reuse it for next cycle [31].
i 7-(3-(4-methoxyphenyl)-1-phenyl-1H-pyrazol-4-yl)-9-methyl-7, 12-dihydro-6Hchromeno[4,3-b] quinoline-6-one (5a)
White solid; yield 86%; mp: 273-275 °C; 1H NMR (300 MHz, CDCl3+DMSO-d6) δ 10.51 (s, 1H), 8.07 (s, 1H), 7.83 (s, 1H), 7.65 (d, J = 8.1 Hz, 2H), 7.52 (d, J = 8.6 Hz, 2H), 7.41 (t, J = 7.7 Hz, 3H), 7.24 (t, J = 7.3 Hz, 1H), 7.01 (d, J = 7.9 Hz, 1H), 6.94 (dd, J = 8.2, 5.4 Hz, 3H), 6.86 (d, J = 8.3 Hz, 1H), 6.79 (t, J = 7.6 Hz, 1H), 6.72 (s, 1H), 5.12 (s, 1H), 3.84 (s, 3H), 2.20 (s, 3H); 13C NMR (75 MHz, CDCl3+DMSO-d6) δ 163.93, 160.48, 158.24, 150.62, 138.29, 135.28, 133.07, 131.00, 130.03, 128.24, 128.09, 127.19, 126.74, 125.64, 124.90, 124.78, 124.15, 119.46, 118.44, 117.87, 116.97, 116.65, 114.53, 112.83, 110.82, 53.95, 34.11, 19.57; ESI-MS: m/z 512 (M + H)+; HRMS (ESI) m/z for C33H26O3N3 calculated m/z: 512.1969, found m/z: 512.1971.
ii 9-methyl-7-(3-(4-nitrophenyl)-1-phenyl-1H-pyrazol-4-yl)-7,12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5b)
Pale yellow solid; yield 82%; mp: 328-330 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.57 (s, 1H), 8.61 (d, J = 3.2 Hz, 2H), 8.25 (d, J = 7.0 Hz, 2H), 8.05 (s, 1H), 7.97 (d, J = 8.2 Hz, 1H), 7.88 (d, J = 8.1 Hz, 1H), 7.83 (s, 1H), 7.74 (s, 2H), 7.58 (d, J = 8.5 Hz, 4H), 7.14 (d, J = 7.8 Hz, 1H), 7.02 (d, J = 5.3 Hz, 2H), 5.17 (s, 1H), 2.20 (s, 3H); 13C NMR (75 MHz, CDCl3+DMSO-d6) δ 163.97, 160.88, 150.62, 148.96, 146.28, 143.06, 138.70, 135.68, 133.36, 131.61, 130.44, 128.85, 128.31, 126.77, 125.89, 124.84, 123.35, 122.37, 121.81, 119.91, 118.19, 117.04, 116.03, 115.00, 112.82, 94.82, 34.19, 19.71; ESI-MS: m/z 527 (M + H)+; HRMS (ESI) m/z for C32H23O4N4 calculated m/z: 527.1714, found m/z: 527.1716.
iii 7-(3-(3,4-dichlorophenyl)-1-phenyl-1H-pyrazol-4-yl)-9-methyl-7, 12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5c)
White solid; yield 76%; mp: 274-276 °C; 1H NMR (300 MHz, CDCl3+DMSO-d6) δ 10.61 (s, 1H), 8.44 (s, 1H), 7.87 (s, 1H), 7.78 (s, 1H), 7.69 (d, J = 6.8 Hz, 2H), 7.57 – 7.49 (m, 2H), 7.44 (d, J = 7.0 Hz, 3H), 7.27 (t, J = 7.2 Hz, 1H), 7.01 (d, J = 7.8 Hz, 1H), 6.92 (d, J = 8.0 Hz, 1H), 6.86 (dd, J = 8.0, 2.2 Hz, 2H), 6.64 (s, 1H), 5.19 (s, 1H), 2.19 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 164.34, 160.41, 148.82, 138.44, 135.64, 133.61, 132.54, 131.31, 131.22, 130.66, 130.58, 129.89, 128.93, 128.59, 127.65, 126.94, 126.82, 126.74, 125.74, 125.15, 120.42, 119.26, 118.33, 117.50, 116.94, 114.93, 34.06, 19.93; ESI-MS: m/z 550 (M + H)+; HRMS (ESI) m/z for C32H22O2N3Cl2 calculated m/z: 550.1084, found m/z: 550.1083.
iv 7-(1,3-diphenyl-1H-pyrazol-4-yl)-9-methyl-7, 12-dihydro-6H-chromeno[4,3b] quinolin-6-one (5d)
Light brown solid; yield 83%; mp: 248-250 °C; 1H NMR (300 MHz, CDCl3+DMSO-d6) δ 10.44 (s, 1H), 7.96 (s, 1H), 7.68 – 7.63 (m, 3H), 7.61 – 7.57 (m, 6H), 7.41 (t, J = 7.4 Hz, 2H), 7.25 (t, J = 7.4 Hz, 1H), 7.02 (d, J = 8.0 Hz, 1H), 6.94 (d, J = 8.0 Hz, 1H), 6.86 (d, J = 8.3 Hz, 1H), 6.78 – 6.71 (m, 2H), 4.96 (s, 1H), 2.21 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 164.21, 161.00, 151.10, 138.60, 135.67, 133.36, 132.13, 131.45, 130.22, 129.40, 128.73, 128.58, 127.40, 127.29, 126.12, 126.02, 125.39, 124.94, 121.20, 119.47, 119.03, 118.26, 117.48, 117.02, 114.89, 34.54, 19.89; ESI-MS: m/z 482 (M + H)+; HRMS (ESI) m/z for C32H24O22N3 calculated m/z: 482.1863, found m/z: 482.1865.
v 7-(3-(4-chlorophenyl)-1-phenyl-1H-pyrazol-4-yl)-9-methyl-7, 12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5e)
White solid; yield 80%; mp: 240-242 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.67 (s, 1H), 8.50 (d, J = 3.2 Hz, 1H), 8.15 (d, J = 7.0 Hz, 1H), 7.94 (d, J = 8.2 Hz, 1H), 7.86 (d, J = 8.2 Hz, 1H), 7.77 (d, J = 8.1 Hz, 1H), 7.72 (s, 1H), 7.63 (s, 2H), 7.48 (d, J = 8.5 Hz, 5H), 7.04 (d, J = 7.8 Hz, 1H), 6.92 (d, J = 5.3 Hz, 3H), 4.96 (s, 1H), 2.20 (s, 3H); 13C NMR (75 MHz, CDCl3+DMSO-d6) δ 164.01, 160.72, 148.80, 146.12, 145.94, 139.74, 138.54, 135.52, 133.20, 131.45, 128.69, 127.78, 126.61, 126.08, 125.34, 124.68, 123.19, 121.65, 119.75, 119.42, 118.03, 117.53, 116.88, 114.84, 112.66, 94.66, 34.03, 19.74; ESI-MS: m/z 516 (M + H)+; HRMS (ESI) m/z for C32H23O2N3Cl calculated m/z: 516.1473, found m/z: 516.1466.
vi 9-methoxy-7-(3-(4-nitrophenyl)-1-phenyl-1H-pyrazol-4-yl)-7, 12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5f)
Yellow solid; yield 84%; mp: 322-324 °C; 1H NMR (300 MHz, CDCl3+DMSO-d6) δ 10.54 (s, 1H), 8.31 – 8.24 (m, 3H), 7.90 (d, J = 8.7 Hz, 2H), 7.84 (d, J = 7.8 Hz, 1H), 7.78 (s, 1H), 7.67 (d, J = 7.9 Hz, 2H), 7.44 (t, J = 7.8 Hz, 3H), 7.29 (t, J = 7.4 Hz, 1H), 6.99 (d, J = 8.6 Hz, 1H), 6.86 (d, J = 8.4 Hz, 1H), 6.77 (dd, J = 8.6, 2.6 Hz, 1H), 6.43 (d, J = 2.0 Hz, 1H), 5.02 (s, 1H), 3.65 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 165.10, 159.18, 153.60, 148.07, 145.39, 137.69, 137.41, 134.64, 129.76, 128.57, 127.75, 126.99, 126.31, 125.86, 125.05, 122.15, 119.84, 118.75, 117.45, 116.64, 115.93, 114.94, 111.90, 110.67, 53.53, 33.19; ESI-MS: m/z 543 (M + H)+; HRMS (ESI) m/z for C32H23O5N4 calculated m/z: 543.1663, found m/z: 543.1665.
vii 7-(3-(3, 4-dichlorophenyl)-1-phenyl-1H-pyrazol-4-yl)-9-methoxy-7, 12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5g)
White solid; yield 81%; mp: 265-267 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.59 (s, 1H), 8.50 (s, 1H), 7.91 (d, J = 7.9 Hz, 1H), 7.80 (s, 1H), 7.70 (d, J = 7.9 Hz, 2H), 7.59 (dd, J = 20.5, 8.4 Hz, 2H), 7.44 (t, J = 7.6 Hz, 3H), 7.28 (t, J = 7.3 Hz, 1H), 6.96 (d, J = 8.6 Hz, 1H), 6.91 – 6.83 (m, 2H), 6.78 (d, J = 8.8 Hz, 1H), 6.33 (s, 1H), 5.13 (s,1H), 3.61 (s, 3H);13C NMR (75 MHz, DMSO-d6) δ 165.10, 159.33, 153.65, 147.90, 137.53, 134.73, 131.65, 130.29, 129.80, 129.15, 128.63, 127.98, 127.78, 126.15, 125.91, 124.93, 119.76, 118.34, 117.49, 116.63, 116.04, 114.97, 111.96, 110.77, 53.60, 33.23; ESI-MS: m/z 566 (M + H)+; HRMS (ESI) m/z for C32H22O3N3Cl2 calculated m/z: 566.1033, found m/z: 566.1035.
viii 9-methoxy-7-(3-(4-methoxyphenyl)-1-phenyl-1H-pyrazol-4-yl)-7, 12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5h)
White solid; yield 88%; mp: 251-253 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.55 (s, 1H), 8.30 (s, 1H), 8.02 (s, 1H), 7.92 (s, 1H), 7.83 (t, J = 8.3 Hz, 2H), 7.67 (t, J = 8.4 Hz, 3H), 7.57 (s, 1H), 7.49 (d, J = 8.4 Hz, 1H), 7.42 – 7.34 (m, 3H), 7.23 (d, J = 4.2 Hz, 1H), 6.99 – 6.95 (m, 2H), 6.86 (s, 1H), 5.16 (s, 1H), 3.83 (s, 3H), 3.62 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 165.13, 160.54, 158.60, 154.80, 151.74, 149.35, 138.63, 135.62, 130.63, 129.38, 128.99, 128.45, 127.87, 126.99, 125.44, 122.48, 120.27, 118.66, 117.24, 116.92, 115.99, 113.25, 112.91, 112.57, 111.54, 93.56, 54.42, 54.30, 34.37; ESI-MS: m/z 528 (M + H)+; HRMS (ESI) m/z for C33H26O4N3 calculated m/z: 528.1918, found m/z: 528.1920.
ix 7-(1,3-diphenyl-1H-pyrazol-4-yl)-9-methoxy-7, 12-dihydro-6H-chromeno[4,3b] quinolin-6-one (5i)
Off white solid; yield 79%; mp: 246-248 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.47 (s, 1H), 8.21 (s, 1H), 7.93 (s, 1H), 7.83 (s, 1H), 7.74 (t, J = 8.3 Hz, 2H), 7.58 (t, J = 8.4 Hz, 3H), 7.47 (d, J = 8.6 Hz, 2H), 7.40 (d, J = 8.4 Hz, 1H), 7.28 (dd, J = 16.2, 6.7 Hz, 4H), 7.14 (d, J = 4.2 Hz, 1H), 6.89 (s, 1H), 6.77 (s, 1H), 5.17 (s, 1H), 3.74 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 165.27, 159.68, 157.74, 153.94, 150.88, 150.16, 148.49, 137.77, 134.76, 129.77, 128.52, 127.65, 126.60, 126.13, 125.25, 124.58, 123.61, 121.62, 119.41, 117.80, 116.38, 115.13, 114.90, 112.39, 110.68, 92.70, 53.44, 33.51; ESI-MS: m/z 498 (M + H)+; HRMS (ESI) m/z for C32H24O3N3 calculated m/z: 498.1812, found m/z: 498.1814.
x 7-(3-(4-chlorophenyl)-1-phenyl-1H-pyrazol-4-yl)-9-methoxy-7, 12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5j)
White solid; yield 76%; mp: 262-264 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.63 (s, 1H), 8.54 (s, 1H), 8.33 (d, J = 8.0 Hz, 1H), 8.27 (d, J = 6.7 Hz, 1H), 7.96 (d, J = 7.5 Hz, 1H), 7.87 (s, 1H), 7.81 (d, J = 7.8 Hz, 2H), 7.68 (s, 2H), 7.47 (d, J = 7.5 Hz, 3H), 7.46 (s, 1H), 7.31 (s, 1H), 6.84 (d, J = 8.5 Hz, 1H), 6.74 (d, J = 9.0 Hz, 1H), 6.33 (d, J = 1.9 Hz, 1H), 5.05 (s, 1H), 3.65 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 164.89, 159.02, 150.47, 148.85, 146.96, 141.91, 137.26, 134.46, 131.25, 129.66, 128.39, 127.60, 127.52, 126.89, 126.29, 125.79, 125.56, 124.55, 121.50, 120.95, 119.74, 117.92, 116.14, 115.71, 114.89, 111.94, 110.50, 92.00, 53.29, 32.84; ESI-MS: m/z 532 (M + H)+; HRMS (ESI) m/z for C32H23O3N3Cl calculated m/z: 532.1425, found m/z: 532.1424.
xi 7-(3-(3,4-dichlorophenyl)-1-phenyl-1H-pyrazol-4-yl)-8,9,10-trimethoxy-7, 12dihydro-6H-chromeno [4,3-b] quinolin-6-one (5k)
White solid; yield 84%; mp: 290-292 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.59 (s, 1H), 8.29 (d, J = 7.9 Hz, 1H), 8.23 – 8.18 (m, 1H), 8.11 (s, 1H), 7.93 (dd, J = 8.3, 1.7 Hz, 1H), 7.76 (d, J = 7.9 Hz, 2H), 7.62 (dd, J = 17.4, 7.9 Hz, 2H), 7.46 – 7.31 (m, 4H), 7.22 (s, 1H), 6.77 (s, 1H), 5.14 (s, 1H), 3.81 (s, 3H), 3.64 (s, 3H), 3.33 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 161.51, 159.96, 151.58, 149.68, 147.62, 145.88, 140.77, 138.52, 136.71, 132.58, 130.41, 128.98, 128.63, 128.44, 127.16, 125.47, 122.67, 122.27, 122.04, 117.57, 116.03, 112.97, 109.68, 95.77, 95.23, 59.63, 58.92, 54.88, 26.02; ESI-MS: m/z 626 (M + H)+; HRMS (ESI) m/z for C34H26O5N3Cl2 calculated m/z: 626.1244, found m/z: 626.1243.
xii 7-(3-(4-chlorophenyl)-1-phenyl-1H-pyrazol-4-yl)-8,9,10-trimethoxy-7, 12-dihydro6H-chromeno [4,3-b] quinolin-6-one (5l)
White solid; yield 90%; mp: 285-287 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.11 (s, 1H), 8.54 (d, J = 7.3 Hz, 1H), 8.28 (d, J = 8.9 Hz, 3H), 7.98 (d, J = 7.8 Hz, 2H), 7.84 (s, 1H), 7.73 – 7.55 (m, 6H), 7.45 (d, J = 6.9 Hz, 1H), 7.01 (s, 1H), 5.23 (s, 1H), 4.05 (s, 3H), 3.87 (s, 3H), 3.50 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 164.01, 159.62, 151.08, 150.69, 148.79, 146.74, 145.00, 141.76, 139.88, 137.63, 135.82, 132.70, 129.52, 128.10, 127.74, 126.27, 124.58, 121.78, 121.38, 116.68, 115.14, 112.08, 108.79, 94.88, 94.35, 58.74, 58.03, 53.99, 25.13; ESI-MS: m/z 592 (M + H)+; HRMS (ESI) m/z for C34H27O5N3Cl calculated m/z: 592.1634, found m/z: 592.1636.
xiii 8,9,10-trimethoxy-7-(3-(4-nitrophenyl)-1-phenyl-1H-pyrazol-4-yl)-7, 12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5m)
Yellow solid; yield 87%; mp: 312-314 °C; 1H NMR (300 MHz, CDCl3+DMSO-d6) δ 10.46 (s, 1H), 8.30 (d, J = 7.9 Hz, 1H), 8.08 (d, J = 7.0 Hz, 2H), 7.91 (d, J = 18.1 Hz, 2H), 7.74 (d, J = 7.9 Hz, 2H), 7.65 (t, J = 7.5 Hz, 1H), 7.50 – 7.43 (m, 4H), 7.38 (d, J = 8.1 Hz, 1H), 7.28 (t, J = 7.3 Hz, 1H), 6.80 (s, 1H), 5.12 (s, 1H), 3.94 (s, 3H), 3.79 (s, 3H), 3.42 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 161.64, 158.09, 152.71, 149.81, 147.75, 146.01, 142.78, 140.90, 138.65, 136.84, 132.24, 130.71, 130.54, 129.11, 128.76, 127.29, 125.60, 122.80, 122.40, 122.17, 117.70, 116.16, 113.10, 109.81, 95.90, 95.36, 59.76, 59.05, 55.01, 34.05; ESI-MS: m/z 603 (M + H)+; HRMS (ESI) m/z for C34H27O7N4 calculated m/z: 603.1874, found m/z: 603.1880.
xiv 8,9,10-trimethoxy-7-(3-(4-methoxyphenyl)-1-phenyl-1H-pyrazol-4-yl)-7,12-dihydro6H-chromeno [4,3-b] quinolin-6-one (5n)
White solid; yield 89%; mp: 309-311 °C; 1H NMR (300 MHz, CDCl3+DMSO-d6) δ 10.53 (s, 1H), 8.71 (d, J = 7.9 Hz, 1H), 8.39 (s, 2H), 8.24 (d, J = 8.0 Hz, 1H), 8.15 (s, 1H), 8.04 (t, J = 7.6 Hz, 1H), 7.77 (dd, J = 16.1, 8.0 Hz, 4H), 7.41 (s, 1H), 7.22 (s, 1H), 7.02 (t, J = 7.5 Hz, 4H), 6.06 (s, 1H), 5.17 (s, 1H) 4.33 – 4.31 (m, 6H), 4.17 (s, 3H), 3.85 (s, 3H); 13C NMR (75 MHz, DMSOd6) δ 165.05, 159.61, 151.07, 150.68, 148.78, 146.73, 144.99, 141.75, 139.87, 137.62, 133.81, 129.69, 129.51, 128.09, 127.73, 127.55, 126.26, 124.57, 121.77, 121.37, 121.14, 116.67, 115.13, 112.07, 108.78, 94.87, 94.34, 58.73, 58.02, 53.98, 34.12; ESI-MS: m/z 588 (M + H)+; HRMS (ESI) m/z for C35H30O6N3 calculated m/z: 588.2129, found m/z: 588.2133.
xv 7-(1,3-diphenyl-1H-pyrazol-4-yl)-8,9,10-trimethoxy-7,12-dihydro-6H-chromeno[4,3b] quinolin-6-one (5o)
Light brown solid; yield 91%; mp: 302-304 °C; 1H NMR (300 MHz, CDCl3+DMSO-d6) δ 10.56 (s, 1H), 8.20 (d, J = 7.9 Hz, 1H), 7.98 (d, J = 7.0 Hz, 2H), 7.81 (d, J = 18.1 Hz, 2H), 7.64 (d, J = 7.9 Hz, 2H), 7.55 (t, J = 7.5 Hz, 1H), 7.40 – 7.31 (m, 5H), 7.28 (d, J = 8.1 Hz, 1H), 7.18 (t, J = 7.3 Hz, 1H), 6.70 (s, 1H), 5.12 (s, 1H), 3.84 (s, 3H), 3.69 (s, 3H), 3.32 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 160.89, 158.08, 152.10, 151.98, 150.51, 142.97, 139.16, 137.15, 134.11, 132.89, 130.61, 129.60, 128.25, 127.23, 126.68, 126.41, 125.24, 122.75, 122.03, 117.69, 116.34, 113.35, 110.53, 96.75, 95.32, 60.01, 59.19, 55.20, 34.22; ESI-MS: m/z 558 (M + H)+; HRMS (ESI) m/z for C34H28O5N3 calculated m/z: 558.2023, found m/z: 558.2028.
xvi 8,10-dimethoxy-7-(3-(4-methoxyphenyl)-1-phenyl-1H-pyrazol-4-yl)-7,12-dihydro6H-chromeno[4,3-b] quinolin-6-one (5p)
White solid; yield 87%; mp: 315-317 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.18 (s, 1H), 8.28 (d, J = 7.9 Hz, 1H), 8.05 (s, 1H), 7.87 (d, J = 8.1 Hz, 2H), 7.69 (d, J = 7.8 Hz, 2H), 7.59 (t, J = 7.4 Hz, 1H), 7.37 (d, J = 7.6 Hz, 4H), 7.15 (d, J = 6.9 Hz, 1H), 6.99 (d, J = 8.1 Hz, 2H), 6.50 (s, 1H), 6.00 (s, 1H), 5.19 (s, 1H), 3.72 (d, J = 21.1 Hz, 6H), 3.12 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 163.51, 159.84, 158.29, 156.97, 151.81, 149.99, 143.29, 138.96, 135.82, 132.80, 131.06, 129.09, 128.84, 128.26, 126.92, 125.20, 123.14, 122.49, 117.37, 116.35, 113.41, 112.89, 105.86, 97.08, 92.82, 55.64, 54.68, 54.26, 34.35; ESI-MS: m/z 558 (M + H)+; HRMS (ESI) m/z for C34H28O5N3 calculated m/z: 558.2023, found m/z: 558.2013.
xvii 7-(1,3-diphenyl-1H-pyrazol-4-yl)-8,10-dimethoxy-7,12-dihydro-6H-chromeno[4,3b] quinolin-6-one (5q)
White solid; yield 81%; mp: 259-261 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.01 (s, 1H), 8.61 (d, J = 7.9 Hz, 1H), 8.38 (s, 1H), 8.20 (d, J = 8.1 Hz, 2H), 8.03 (d, J = 7.8 Hz, 2H), 7.92 (t, J = 7.4 Hz, 1H), 7.70 (d, J = 7.6 Hz, 4H), 7.49 (d, J = 6.9 Hz, 2H), 7.32 (d, J = 8.1 Hz, 2H), 6.84 (s, 1H), 6.34 (s, 1H), 5.23 (s, 1H), 3.64 (s, 3H), 3.46 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 163.14, 158.57, 157.57, 154.46, 146.76, 136.65, 135.93, 133.55, 130.91, 129.32, 128.60, 127.58, 127.41, 127.05, 126.69, 126.49, 126.13, 125.18, 124.60, 123.75, 123.45, 120.21, 116.25, 116.01, 115.81, 115.38, 101.43, 91.41, 90.35, 52.81, 52.47, 33.44; ESI-MS: m/z 528 (M + H)+; HRMS (ESI) m/z for C33H26O4N3 calculated m/z: 528.1918, found m/z: 528.1906.
xviii 7-(3-(3,4-dichlorophenyl)-1-phenyl-1H-pyrazol-4-yl)-8,10-dimethoxy-7,12-dihydro6H-chromeno [4,3-b] quinolin-6-one (5r)
White solid; yield 78%; mp: 315-317 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.19 (s, 1H), 8.29 (d, J = 7.9 Hz, 1H), 8.23 – 8.19 (m, 1H), 8.11 (s, 1H), 7.93 (dd, J = 8.3, 1.7 Hz, 1H), 7.76 (d, J = 7.9 Hz, 2H), 7.62 (dd, J = 17.4, 7.9 Hz, 2H), 7.47 – 7.28 (m, 5H), 7.22 (s, 1H), 6.77 (s, 1H), 5.26 (s, 1H), 3.81 (s, 3H), 3.64 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 163.70, 159.14, 158.13, 155.02, 147.33, 137.22, 136.50, 134.12, 131.48, 129.17, 128.14, 127.98, 127.61, 127.06, 126.70, 125.75, 125.17, 124.02, 120.78, 116.81, 115.94, 102.00, 91.98, 90.92, 53.37, 53.04, 34.71; ESIMS: m/z 596 (M + H)+; HRMS (ESI) m/z for C33H24O4N3Cl2 calculated m/z: 596.1138, found m/z: 596.1140.
xix 7-(3-(4-chlorophenyl)-1-phenyl-1H-pyrazol-4-yl)-8,10-dimethoxy-7,12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5s)
White solid; yield 81%; mp: 278-280 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.30 (s, 1H), 8.31 (s, 1H), 8.11 (d, J = 4.1 Hz, 2H), 8.06 (d, J = 11.2 Hz, 2H), 7.80 (d, J = 8.5 Hz, 1H), 7.68 (d, J = 8.6 Hz, 2H), 7.62 (s, 1H), 7.53 (d, J = 8.1 Hz, 2H), 7.40 (t, J = 7.7 Hz, 1H), 7.25 (d, J = 8.3 Hz, 2H), 6.47 (s, 1H), 6.18 (s, 1H), 5.08 (s, 1H), 3.56 (d, J = 4.6 Hz, 6H); 13C NMR (75 MHz, DMSO) δ 163.93, 159.36, 158.36, 157.61, 155.25, 147.56, 137.45, 136.72, 134.35, 131.70, 130.11, 129.40, 128.37, 128.20, 127.48, 127.28, 126.92, 125.98, 124.54, 124.24, 121.01, 117.04, 116.81, 116.61, 114.94, 102.22, 97.16, 92.20, 91.14, 53.60, 53.26, 34.28; ESI-MS: m/z 562 (M + H)+; HRMS (ESI) m/z for C33H25O4N3Cl calculated m/z: 562.1528, found m/z: 562.1519. 9505984890.
xx 8,10-dimethoxy-7-(3-(4-nitrophenyl)-1-phenyl-1H-pyrazol-4-yl)-7,12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5t)
Pale yellow solid; yield 85%; mp: 293-295 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.51 (s, 1H), 8.51 (s, 1H), 8.31 (d, J = 4.1 Hz, 2H), 8.24 (s, 2H), 8.00 (d, J = 8.5 Hz, 1H), 7.89 (d, J = 8.6 Hz, 2H), 7.81 (d, J = 8.7 Hz, 1H), 7.73 (d, J = 8.1 Hz, 2H), 7.60 (t, J = 7.7 Hz, 1H), 7.46 (d, J = 8.3 Hz, 2H), 6.68 (s, 1H), 6.38 (s, 1H), 5.28 (s, 1H), 3.76 (d, J = 4.6 Hz, 6H); 13C NMR (75 MHz, DMSO-d6) δ 163.67, 157.99, 157.61, 155.21, 154.91, 150.22, 146.12, 144.85, 136.96, 136.35, 133.28, 129.15, 128.12, 127.21, 126.99, 125.70, 124.38, 123.97, 121.45, 120.87, 120.73, 117.98, 116.43, 116.09, 114.61, 103.86, 101.99, 95.36, 91.90, 90.77, 55.80, 53.23, 34.72; ESI-MS: m/z 573 (M + H)+; HRMS (ESI) m/z for C32H25O4N3Cl calculated m/z: 573.1768, found m/z: 573.1767.
xxi 8,10-dimethoxy-7-(3-(4-methoxyphenyl)-1-phenyl-1H-pyrazol-4-yl)-7,12-dihydro6H-chromeno[4,3-b] quinolin-6-one (5u)
White solid; yield 88%; mp: 220-222 °C; 1H NMR (300 MHz, CDCl3+DMSO-d6) δ 10.54 (s, 1H), 8.51 (s, 1H), 8.23 (d, J = 8.0 Hz, 1H), 7.78 (d, J = 8.5 Hz, 2H), 7.61 (d, J = 18.6 Hz, 3H), 7.35 (t, J = 8.4 Hz, 4H), 7.18 (t, J = 7.3 Hz, 1H), 7.03 (d, J = 7.1 Hz, 2H), 6.88 (s, 1H), 6.16 (s, 1H), 5.27 (s, 1H), 3.86 (s, 3H), 3.79 (s, 3H), 3.45 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 163.62, 159.61, 157.52, 150.83, 148.54, 146.11, 143.53, 142.18, 137.87, 129.36, 128.32, 127.32, 126.30, 124.69, 124.16, 123.91, 121.51, 120.79, 116.33, 115.15, 114.77, 112.10, 109.30, 99.04, 93.14, 54.97, 53.86, 53.67, 34.71; ESI-MS: m/z 558 (M + H)+; HRMS (ESI) m/z for C34H28O5N3 calculated m/z: 558.2024, found m/z: 558.201.
xxii 8,10-dimethoxy-7-(3-(4-nitrophenyl)-1-phenyl-1H-pyrazol-4-yl)-7,12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5v)
White solid; yield 81%; mp: 291-293 °C; 1H NMR (300 MHz, CDCl3+DMSO-d6) δ 10.58 (s, 1H), 8.48 (s, 1H), 8.23 (d, J = 7.9 Hz, 1H), 8.00 (d, J = 7.0 Hz, 2H), 7.83 (d, J = 18.1 Hz, 2H), 7.66 (d, J = 7.9 Hz, 2H), 7.58 (t, J = 7.5 Hz, 1H), 7.40 (dd, J = 14.4, 7.4 Hz, 4H), 7.30 (d, J = 8.1 Hz, 1H), 7.21 (t, J = 7.3 Hz, 1H), 6.73 (s, 1H), 5.24 (s, 1H), 3.86 (s, 3H), 3.71 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 164.69, 158.17, 147.46, 144.91, 142.42, 136.96, 134.00, 129.16, 127.51, 127.42, 127.17, 126.63, 125.94, 121.79, 121.33, 119.66, 118.90, 117.04, 116.20, 114.77, 54.11, 53.64, 34.35; ESI-MS: m/z 573 (M + H)+; HRMS (ESI) m/z for C33H25O6N4 calculated m/z: 573.1769, found m/z: 573.1771.
xxiii 7-(1,3-diphenyl-1H-pyrazol-4-yl)-9,10-dimethoxy-7,12-dihydro-6H-chromeno[4,3b] quinolin-6-one (5w)
White solid; yield 78%; mp: 227-229 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.57 (s, 1H), 8.58 (d, J = 7.9 Hz, 1H), 8.34 (s, 1H), 8.17 (d, J = 8.1 Hz, 2H), 7.99 (d, J = 7.8 Hz, 2H), 7.89 (t, J = 7.4 Hz, 1H), 7.67 (dd, J = 15.9, 7.6 Hz, 4H), 7.45 (d, J = 6.9 Hz, 2H), 7.28 (d, J = 8.1 Hz, 2H), 6.80 (s, 1H), 6.30 (s, 1H), 5.29 (s, 1H), 3.62 (s, 3H), 3.42 (s, 3H); 13C NMR (75 MHz, DMSOd6) δ 163.68, 159.12, 158.11, 157.36, 155.00, 147.31, 137.20, 136.48, 134.10, 131.46, 129.86, 129.15, 128.12, 127.96, 127.59, 126.68, 125.73, 124.30, 124.00, 120.76, 116.56, 116.36, 115.92, 101.98, 91.96, 90.90, 53.35, 53.02, 34.29; ESI-MS: m/z 528 (M + H)+; HRMS (ESI) m/z for C33H26O4N3 calculated m/z: 528.1918, found m/z: 528.1917.
xxiv 7-(3-(4-chlorophenyl)-1-phenyl-1H-pyrazol-4-yl)-9,10-dimethoxy-7,12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5x)
White solid; yield 80%; mp: 236-238 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.32 (s, 1H), 8.34 (s, 1H), 8.11 (d, J = 4.1 Hz, 2H), 8.10 (d, J = 11.2 Hz, 2H), 7.80 (d, J = 8.5 Hz, 1H), 7.64 (d, J = 8.6 Hz, 2H), 7.62 (s, 1H), 7.43 (d, J = 8.1 Hz, 1H), 7.38 (t, J = 7.7 Hz, 2H), 7.29 (d, J = 8.3 Hz, 2H), 6.43 (s, 1H), 6.22 (s, 1H), 5.10 (s, 1H), 3.62 (s, 3H), 3.56 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 163.56, 158.35, 150.22, 149.81, 147.53, 145.47, 140.50, 138.61, 136.37, 134.55, 132.01, 128.43, 128.25, 126.83, 126.48, 125.00, 123.31, 120.52, 120.11, 119.89, 115.42, 113.87, 110.81, 107.52, 93.61, 93.08, 57.47, 56.76, 33.56; ESI-MS: m/z 562 (M + H)+; HRMS (ESI) m/z for C33H25O4N3Cl calculated m/z: 562.1528, found m/z: 562.1525.
xxv 7-(3-(3,4-dichlorophenyl)-1-phenyl-1H-pyrazol-4-yl)-9,10-dimethoxy-7,12-dihydro6H-chromeno [4,3-b] quinolin-6-one (5y)
White solid; yield 78%; mp: 212-214 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.38 (s, 1H), 8.38 (d, J = 7.9 Hz, 1H), 8.32 – 8.28 (m, 1H), 8.20 (s, 1H), 8.02 (dd, J = 8.3, 1.7 Hz, 1H), 7.85 (d, J = 7.9 Hz, 2H), 7.72 (dd, J = 17.4, 7.9 Hz, 2H), 7.56 – 7.38 (m, 4H), 7.31 (s, 1H), 6.87 (s, 2H), 5.13 (s, 1H), 3.91 (s, 3H), 3.73 (s, 3H); 13C NMR (75 MHz, DMSO) δ 163.18, 151.08, 150.69, 148.79, 146.74, 145.00, 141.76, 139.88, 137.63, 135.82, 129.70, 129.52, 128.10, 127.74, 127.56, 126.27, 124.58, 121.78, 121.38, 121.15, 116.68, 115.14, 112.08, 108.79, 94.88, 94.35, 58.74, 58.03, 34.30; ESI-MS: m/z 596 (M + H)+; HRMS (ESI) m/z for C33H24O4N3Cl2 calculated m/z: 596.1138, found m/z: 596.1130.
xxvi 9-chloro-7-(1,3-diphenyl-1H-pyrazol-4-yl)-7,12-dihydro-6H-chromeno[4,3b] quinolin-6-one (5z)
White solid; yield 78%; mp: 287-289 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.77 (s, 1H), 8.25 (s, 1H), 8.05 (d, J = 8.2 Hz, 1H), 7.97 (d, J = 8.2 Hz, 1H), 7.88 (d, J = 8.1 Hz, 2H), 7.83 (s, 1H), 7.66 (d, J = 8.5 Hz, 4H), 7.58-7.50 (m, 5H), 7.14 (d, J = 7.8 Hz, 1H), 7.02 (d, J = 5.3 Hz, 3H), 5.17 (s, 1H); 13C NMR (75 MHz, DMSO-d6) δ 163.23, 153.92, 149.19, 147.30, 134.80, 131.59, 130.00, 129.88, 128.73, 127.94, 126.98, 125.90, 125.57, 124.88, 121.84, 121.29, 120.08, 118.26, 117.64, 116.48, 116.05, 115.23, 114.93, 112.28, 111.96, 110.84, 92.33, 33.28; ESI-MS: m/z 502 (M + H)+; HRMS (ESI) m/z for C31H21O2N3Cl calculated m/z: 502.1317, found m/z: 502.1319.
xxvii 9-chloro-7-(3-(4-methoxyphenyl)-1-phenyl-1H-pyrazol-4-yl)-7,12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5aa)
White solid; yield 82%; mp: 292-294 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.60 (s, 1H), 8.51 (s, 1H), 8.30 (d, J = 8.0 Hz, 1H), 8.24 (d, J = 6.7 Hz, 1H), 7.92 (d, J = 7.5 Hz, 1H), 7.84 (s, 1H), 7.78 (d, J = 7.8 Hz, 1H), 7.65 (d, J = 7.6 Hz, 2H), 7.44 (d, J = 7.5 Hz, 2H), 7.43 (d, J = 7.5 Hz, 2H), 7.29 (d, J = 7.2 Hz, 2H), 6.81 (d, J = 8.5 Hz, 1H), 6.71 (d, J = 9.0 Hz, 1H), 6.30 (d, J = 1.9 Hz, 1H), 5.34 (s, 1H), 3.59 (s, 3H); 13C NMR (75 MHz, CDCl3+DMSO-d6) δ 163.01, 159.23, 152.92, 148.80, 146.12, 145.94, 138.54, 133.20, 132.04, 130.28, 128.69, 127.78, 126.61, 125.73, 124.68, 123.19, 122.68, 121.65, 119.75, 119.42, 118.03, 117.53, 116.88, 114.84, 112.66, 94.66, 52.46, 34.43; ESI-MS: m/z 532 (M + H)+; HRMS (ESI) m/z for C32H23O3N3Cl calculated m/z: 532.1422, found m/z: 532.1420.
xxviii 9-chloro-7-(3-(4-nitrophenyl)-1-phenyl-1H-pyrazol-4-yl)-7,12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5ab)
White solid; yield 72%; mp: 246-248 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.51 (s, 1H), 8.83 (d, J = 7.3 Hz, 1H), 8.38 (d, J = 8.9 Hz, 2H), 8.08 (d, J = 7.8 Hz, 2H), 7.94 (s, 1H), 7.83 – 7.65 (m, 6H), 7.54 (s, 2H), 7.36 (d, J = 8.1 Hz, 2H), 6.21 (d, J = 7.6 Hz, 1H), 5.35 (s, 1H); 13C NMR (75 MHz, DMSO-d6) δ 163.23, 152.44, 151.32, 149.84, 149.53, 142.31, 138.50, 136.49, 133.45, 130.23, 129.95,128.48, 128.22, 127.94, 127.59, 126.57, 126.02, 125.75, 124.58, 122.09, 121.37, 117.03, 115.68, 112.69, 109.87, 96.09, 94.66, 34.56; ESI-MS: m/z 547 (M + H)+; HRMS (ESI) m/z for C31H20O4N4Cl calculated m/z: 547.1168, found m/z: 547.1169.
xxix 9-chloro-7-(3-(4-chlorophenyl)-1-phenyl-1H-pyrazol-4-yl)-7,12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5ac)
White solid; yield 74%; mp: 286-288 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.53 (s, 1H), 8.14 (s, 1H), 7.94 (d, J = 4.1 Hz, 2H), 7.89 (d, J = 11.2 Hz, 2H), 7.63 (d, J = 8.5 Hz, 2H), 7.51 (d, J = 8.6 Hz, 2H), 7.45 (s, 1H), 7.36 (d, J = 8.1 Hz, 2H), 7.23 (t, J = 7.7 Hz, 1H), 7.08 (d, J = 8.3 Hz, 2H), 6.30 (s, 1H), 6.01 (d, J = 7.6 Hz, 1H), 5.27 (s, 1H); 13C NMR (75 MHz, DMSO-d6) δ 164.93, 158.61, 152.25, 148.56, 138.45, 137.72, 135.35, 132.70, 131.11, 130.40, 129.37, 129.20, 128.84, 127.92, 126.98, 126.40, 125.54, 125.24, 122.01, 118.04, 117.81, 117.61, 117.17, 103.22, 93.20, 92.14, 34.24; ESI-MS: m/z 536 (M + H)+; HRMS (ESI) m/z for C31H20O2N3 Cl2 calculated m/z: 536.0927, found m/z: 536.0929.
xxx 9-chloro-7-(3-(3,4-dichlorophenyl)-1-phenyl-1H-pyrazol-4-yl)-7,12-dihydro-6Hchromeno[4,3-b] quinolin-6-one (5ad)
White solid; yield 70%; mp: 230-232 °C; 1H NMR (300 MHz, DMSO-d6) δ 10.55 (s, 1H), 8.46 (s, 1H), 8.26 (d, J = 4.1 Hz, 2H), 8.20 (d, J = 11.2 Hz, 2H), 7.95 (d, J = 8.5 Hz, 1H), 7.83 (d, J = 8.6 Hz, 2H), 7.77 (s, 1H), 7.68 (d, J = 8.1 Hz, 2H), 7.54 (t, J = 7.7 Hz, 1H), 7.40 (d, J = 8.3 Hz, 2H), 6.62 (s, 1H), 6.32 (s, 1H), 5.22 (s, 1H); 13C NMR (75 MHz, DMSO-d6) δ 164.56, 158.99, 152.88, 148.18, 138.07, 137.35, 134.97, 132.33, 130.74, 129.00, 128.83, 128.47, 127.91, 127.55, 126.61, 126.02, 125.17, 124.87, 121.63, 117.67, 117.44, 117.23, 116.80, 102.85, 92.83, 91.77, 34.57; ESI-MS: m/z 570 (M + H)+; HRMS (ESI) m/z for C31H19O2N3Cl3 calculated m/z: 570.0537, found m/z: 570.0539.
IV Biology
i Cytotoxic Activity
The anticancer activities of the complexes were determined using MTT assay [32]. Cancer cells (DU-145, MCF-7, HeLa and A549) were used in this assay. 1×104 cells/well were seeded in 200 µL Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% FBS in each well of 96-well micro-culture plates and incubated for 24 h at 37 oC in a CO2 incubator. All the derivatives diluted to the desired concentrations (500 nM, 1 µM, 5 µM, 10 µM, 25 µM, 50 µM, 75 µM, 100 µM, 150 µM) in culture medium, were added to the wells with respective vehicle control. Doxorubicin treated cells in the same concentration range were used as control. After 48 h of incubation, 10 µL MTT (3-(4,5-dimethylthiazol-2-yl) - 2,5-diphenyl tetrazolium bromide) (5 mg/mL) was added to each well and the plates were further incubated for 4 h. Then the supernatant from each well was carefully removed, formazan crystals were dissolved in 100 µL of DMSO and absorbance at 570 nm wavelength was recorded at a wavelength of 540 nm using an ELx800 microplate reader (BioTek, USA).
ii UV-Visible Studies
UV-visible spectroscopy titrations were performed using ABI Lambda 40 UV-Vis spectrophotometer (Foster City, USA) at 25 oC using 1 cm path length quartz cuvette. Stock solutions of 1 mM of CT DNA (calf thymus DNA, which can form perfect double stranded DNA structure) were prepared in 100 mM Tris-HCl (pH 7.0). 1 mM stock solution of the synthesized 5s and 5t compounds was prepared by dissolving them in 1:1 DMSO: Milli Q water. UV-visible absorption titrations were performed by adding 10 µM CT DNA solution in 100 mM Tris-HCl (pH 7.0) each time to the quartz cuvette containing about 10 µM derivative solution. Titrations were carried out until the complex absorption band remained at a fixed wavelength upon five successive additions of CT DNA. Absorption spectra were recorded from 200 nm to 400 nm.
iii Fluorescence Studies
Fluorescence emission spectra were measured at 25 oC using a Hitachi F7000 spectrofluorimeter (Maryland, USA) using a 1 cm path length quartz cuvette. Throughout the fluorescence experiment, the concentration of the 5s and 5t compounds were kept constant (10 µM) and titrated with increasing concentrations of CT DNA (each addition with an increment of 10 µM CT DNA). Fluorescence spectra were recorded after each addition of CT DNA to the fluorescent cuvette. After each experiment, the quartz cuvettes was thoroughly washed with distilled water and dilute nitric acid (approximately 0.1 N, nitric acid) to remove traces of derivative binding to the walls of quartz cuvette. 5s and 5t derivatives were excited at 280 nm and 290 nm, respectively, and emission spectra for each titration were collected in the range from 270 nm to 300 nm. Each spectrum was recorded three times and the average of three scans was taken.
iv CD Studies
Circular dichroism experiments were carried out using JASCO 815 CD spectropolarimeter (Jasco, Tokyo, Japan). CD spectrum was recorded from 230 to 300 nm to find the conformation of DNA after CT DNA-compound interaction. For CD experiments, 10 × 10-6 M of CT DNA was used. For characterizing compound-CT DNA interaction, CD spectra was recorded in 1:0, 1:1 and 1:2 molar ratio of CT DNA-compound, respectively. CD titrations were performed in 100 mM Tris-HCl (pH 7.0) at 25 oC. Each CD spectrum was recorded thrice and the average of three scans was considered.
v Viscosity Studies
Viscosity experiments were conducted using Ostwald viscometer immersed in a water bath and maintained at 25 oC. Viscosity experiments were performed for each compound (15 µM), after mixing them with CT-DNA solution (150 µM). Before mixing DNA and compounds, viscosity measurements were performed with CT-DNA alone. EtBr-CT-DNA complexes were considered as control. DNA solution was prepared in 100 mM Tris-HCl (pH 7.0). Graph was drawn by plotting (ŋ/ŋo)1/3 versus compound/CT-DNA, where ŋ is the viscosity of CT-DNA in the presence of compounds and ŋo is the viscosity of CT-DNA alone. Viscosity values were calculated according to the protocol mentioned by Tan et al. [33].
vi Topoisomerase Inhibition Studies
Topoisomerase I inhibitory activity was measured using a DNA cleavage assay as described previously [34]. pBR322 plasmid DNA was purchased from Sigma Aldrich, USA and 0.5 µg of pBR322 DNA was incubated with 1 unit of Topo I enzyme (Invitrogen) in 1X NEB buffer (50 mM potassium acetate, 20 mM Tris acetate, 10 mM magnesium acetate, 1 mM DTT). Both the synthetic derivatives (5s and 5t) at 20 µM were added to the Topo I-DNA complex and incubated at 37 oC for 30 min, allowing the formation of ternary enzyme-DNA-ligand complex. Camptothecin was used as a control. Then the enzyme was inactivated by increasing the temperature to 65 oC. After the incubation, the samples were resolved using 1% agarose gel electrophoresis which enabled the visualization of cleavage products. The pBR322 DNA sample to which no ligand was added acted as positive control.
Acknowledgements
Authors J.K, B.N and S.R.R are thankful to Department of Science & Technology, New Delhi for providing fellowships under DST-INSPIRE Fellowship programme and CSIR-IICT-Hyderabad. We also extend our appreciation to the International Scientific Partnership Program ISPP at King Saud University for funding this research work through ISPP#0054 and Vardhaman College of Engineering for providing support.
Article Info
Article Type
Research ArticlePublication history
Received: Sun 01, Mar 2020Accepted: Mon 13, Apr 2020
Published: Thu 30, Apr 2020
Copyright
© 2023 C. Ganesh Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.AJMC.2020.01.05
Author Info
Ahmed Kamal Burri Nagaraju C. Ganesh Kumar Jeshma Kovvuri Jitendra Gour Kishore Mullagiri Narayana Nagesh Sunitha Rani Routhu
Corresponding Author
C. Ganesh KumarOrganic Synthesis and Process Chemistry Division, CSIR-Indian Institute of Chemical Technology, Hyderabad, Telangana, India
Figures & Tables

|
Entry |
Catalyst |
Temperature (°C) |
Time (h) |
Solvent |
Yielda (%) |
|
1 |
p-TSA |
80 |
8 |
EtOH |
20 |
|
2 |
p-TSA |
100 |
8 |
H2O |
10 |
|
3 |
L-proline |
100 |
8 |
EtOH |
30 |
|
4 |
AcOH |
80 |
8 |
EtOH |
53 |
|
5 |
NH2SO3H |
80 |
8 |
EtOH |
65 |
|
6 |
NH2SO3H |
100 |
8 |
EtOH |
49 |
|
7 |
Amberlite IR-120 H |
80 |
5 |
EtOH |
86 |
|
8 |
Amberlite IR-120 H |
100 |
5 |
H2O |
32 |
|
9 |
Amberlite IR-120 H |
120 |
8 |
DMF |
68 |
|
10 |
Amberlite IR-120 H |
40 |
5 |
DCM |
30 |
|
11 |
Amberlite IR-120 H |
60 |
5 |
CHCl3 |
55 |
|
12 |
Amberlite IR-120 H |
80 |
5 |
ACN |
60 |
|
13 |
Amberlite IR-120 H |
85 |
8 |
EtOH-H2O |
51 |
|
14 |
Amberlite IR-120 H |
rt |
8 |
EtOH |
30 |
|
15 |
-- |
rt |
12 |
-- |
-- |
|
16 |
-- |
100 |
12 |
-- |
15 |
|
17 |
Amberlite IR-120 H |
100 |
12 |
-- |
trace |
aIsolated yields
Table 2: Cytotoxicity of 1,3-diphenylpyrazole-chromenoquinolin-6-one compounds (5a‒ad) on selected human cancer cell lines.
|
|
Compound |
|
|
|
|
|
IC50 values (µM) a |
|
|
|
|
||||
|
|
|
|
A549b |
|
|
MCF-7c |
|
|
HeLad |
|
|
DU-145e |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
5a |
21.26 ± 1.08 |
|
9.91 ± 0.55 |
|
22.38 ± 1.55 |
|
18.32 ± 0.88 |
|
|
|||||
|
|
5b |
26.91 ± 1.92 |
|
8.20 ± 0.42 |
|
22.96 ± 1.32 |
|
17.51 ± 0.92 |
|
|
|||||
|
|
5c |
32.35 ± 1.86 |
|
10.73± 0.34 |
|
39.90 ± 2.22 |
|
21.85 ± 0.86 |
|
|
|||||
|
|
5d |
50.11 ± 2.78 |
|
15.27 ± 0.46 |
|
62.57 ± 3.96 |
|
30.85 ± 1.78 |
|
|
|||||
|
|
5e |
6.46 ± 0.99 |
|
5.46 ± 0.51 |
|
14.79 ± 2.02 |
|
6.31 ± 0.99 |
|
|
|||||
|
|
5f |
10.81 ± 0.63 |
|
1.98 ± 0.71 |
|
3.23 ± 0.22 |
|
4.69 ± 0.63 |
|
|
|||||
|
|
5g |
11.32 ± 0.89 |
|
3.68 ± 0.91 |
|
14.12 ± 1.02 |
|
6.53 ± 0.89 |
|
|
|||||
|
|
5h |
25.50 ± 1.54 |
|
11.70 ± 0.61 |
|
25.11 ± 1.86 |
|
18.01 ± 1.54 |
|
|
|||||
|
|
5i |
19.66 ± 0.98 |
|
5.02 ± 0.71 |
|
18.38 ± 1.04 |
|
10.78 ± 0.98 |
|
|
|||||
|
|
5j |
15.55 ± 1.52 |
|
1.82 ± 0.24 |
|
4.68 ± 0.31 |
|
6.84 ± 0.52 |
|
|
|||||
|
|
5k |
15.24 ± 1.26 |
|
4.68 ± 0.46 |
|
18.72 ± 1.02 |
|
9.14± 1.26 |
|
|
|||||
|
|
5l |
16.98 ± 0.52 |
|
3.65 ± 0.23 |
|
15.70 ± 1.03 |
|
6.61 ± 0.52 |
|
|
|||||
|
|
5m |
16.73 ± 1.06 |
|
3.50 ± 0.96 |
|
7.24 ± 0.98 |
|
10.47 ± 1.86 |
|
|
|||||
|
|
5n |
39.81 ± 2.05 |
|
12.56 ± 1.66 |
|
19.67 ±1.96 |
|
21.92 ± 1.26 |
|
|
|||||
|
|
5o |
24.83 ± 1.52 |
|
17.17 ± 1.03 |
|
19.61 ± 2.02 |
|
20.18 ± 0.64 |
|
|
|||||
|
|
5p |
14.21 ± 1.29 |
|
11.81 ± 0.94 |
|
21.72 ± 1.03 |
|
11.22 ± 1.69 |
|
|
|||||
|
|
5q |
13.30 ± 1.38 |
|
2.75 ± 0.54 |
|
4.26 ± 0.76 |
|
6.18 ± 0.96 |
|
|
|||||
|
|
5r |
15.88 ± 1.62 |
|
2.80 ± 0.84 |
|
4.36 ± 0.12 |
|
6.92 ± 0.47 |
|
|
|||||
|
|
5s |
10.59 ± 0.43 |
|
1.22 ± 0.94 |
|
3.51 ± 0.13 |
|
2.99 ± 0.13 |
|
|
|||||
|
|
5t |
5.25 ± 0.23 |
|
1.64 ± 0.76 |
|
2.71 ± 0.33 |
|
4.52 ± 0.37 |
|
|
|||||
|
|
5u |
19.05 ± 2.69 |
|
11.41 ± 0.52 |
|
17.37 ± 0.02 |
|
13.58 ± 2.69 |
|
|
|||||
|
|
5v |
17.11 ± 1.36 |
|
3.23 ± 0.98 |
|
10.56 ± 0.41 |
|
7.49 ± 0.76 |
|
|
|||||
|
|
5w |
21.00 ± 1.65 |
|
9.75 ± 0.33 |
|
16.18 ± 0.66 |
|
15.89 ± 1.65 |
|
|
|||||
|
|
5x |
15.88 ± 1.20 |
|
2.81 ± 0.87 |
|
4.36 ± 0.53 |
|
6.92 ± 0.60 |
|
|
|||||
|
|
5y |
11.35 ± 1.23 |
|
9.53 ± 0.91 |
|
23.77± 1.89 |
|
10.30 ± 1.23 |
|
|
|||||
|
|
5z |
26.30 ± 2.36 |
|
13.92 ± 0.78 |
|
24.85 ± 2.49 |
|
19.56 ± 1.36 |
|
|
|||||
|
|
5aa |
28.84 ± 2.13 |
|
20.14 ± 1.02 |
|
26.91 ± 1.26 |
|
22.40 ± 1.73 |
|
|
|||||
|
|
5ab |
17.37 ± 0.95 |
|
12.73 ± 0.51 |
|
16.87 ± 1.23 |
|
14.12 ± 0.61 |
|
|
|||||
|
|
5ac |
17.78 ± 1.05 |
|
13.66 ± 1.56 |
|
21.18 ± 2.01 |
|
17.00 ± 1.05 |
|
|
|||||
|
|
5ad |
20.41 ± 0.92 |
|
12.30 ± 1.32 |
|
20.46 ± 1.36 |
|
18.31 ± 0.76 |
|
|
|||||
|
|
Doxorubicin |
1.24 ± 0.59 |
|
1.32 ± 0.11 |
|
1.53 ± 0.47 |
|
2.11 ± 0.70 |
|
|
|||||
|
|
(Positive control) |
|
|
|
|
|
|||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
a 50% Inhibitory concentration after 48 h of drug treatment and the values are average of three individual experiments; bHuman lung cancer; cHuman breast cancer; dHuman cervical cancer; dHuman prostate cancer.






References
- Bray F, Jemal A, Grey N, Ferlay J, Forman D (2012) Global cancer transitions according to the Human Development Index (2008-2030): a population-based study. Lancet Oncol 13: 790-801. [Crossref]; b) Jemal A, Bray F, Center MM, Ferlay J, Ward E et al. (2011) Global cancer statistics. CA Cancer J Clin 61: 69-90. [Crossref]
- Varmus H (2006) The new era in cancer research. Science 312: 1162-1165. [Crossref]; b) Vineis P, Wild CP (2014) Global cancer patterns: causes and prevention. Lancet 383: 549-557. [Crossref]
- C Bourdouxhe-Housiaux, P Colson, C Houssier, MJ Waring, C Bailly (1996) Biochemistry 35: 4251-4264; b) Sheng J, Gan J, Huang Z (2013) Structure-based DNA-targeting strategies with small molecule ligands for drug discovery. Med Res Rev 33: 1119-1173. [Crossref]
- Lu Y, Ran T, Lin G, Jin Q, Jin J et al. (2014) Novel 1H-pyrazole-3-carboxamide derivatives: synthesis, anticancer evaluation and identification of their DNA-binding interaction. Chem Pharm Bull 62: 238-246. [Crossref]
- a) Keter FK, Darkwa J (2012) Perspective: the potential of pyrazole-based compounds in medicine. Biometals 25: 9-21. [Crossref]; b) Kumar V, Kaur K, Gupta GK, Sharma AK (2013) Pyrazole containing natural products: synthetic preview and biological significance. Eur J Med Chem 69: 735-753. [Crossref]
- B Caliskan, A Yilmaz, I Evren, S Menevse, O Uludag et al. (2013) Med Chem Res 22: 782-793.
- Gilbert AM, Failli A, Shumsky J, Yang Y, Severin A et al. (2006) Pyrazolidine-3,5-diones and 5-hydroxy-1H-pyrazol-3(2H)-ones, inhibitors of UDP-N-acetylenolpyruvyl glucosamine reductase. J Med Chem 49: 6027-6036. [Crossref]
- Abdel-Aziz M, Abuo-Rahma Gel-D, Hassan AA (2009) Synthesis of novel pyrazole derivatives and evaluation of their antidepressant and anticonvulsant activities. Eur J Med Chem 44: 3480- 3487. [Crossref]
- Shih SR, Chu TY, Reddy GR, Tseng SN, Chen HL (2010) Pyrazole compound BPR1P0034 with potent and selective anti-influenza virus activity. J Biomed Sci 17: 13. [Crossref]
- a) Kumar H, Saini D, Jain S, Jain N (2013) Pyrazole scaffold: a remarkable tool in the development of anticancer agents. Eur J Med Chem 70: 248-258. [Crossref]
- a) Baviskar AT, Banerjee UC, Gupta M, Singh R, Kumar S et al. (2013) Synthesis of imine-pyrazolopyrimidinones and their mechanistic interventions on anticancer activity. Bioorg Med Chem 21: 5782-5793. [Crossref]; b) Tao XX, Duan YT, Chen LW, Tang DJ, Yang MR et al. (2016) Design, synthesis and biological evaluation of pyrazolyl-nitroimidazole derivatives as potential EGFR/HER-2 kinase inhibitors. Bioorg Med Chem Lett 26: 677-683. [Crossref]; c) Miyamoto N, Sakai N, Hirayama T, Miwa K, Oguro Y et al. (2013) Discovery of N-[5-({2-[(cyclopropylcarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}oxy)-2-methylphenyl]-1,3-dimethyl-1H-pyrazole-5-carboxamide (TAK-593), a highly potent VEGFR2 kinase inhibitor. Bioorg Med Chem 21: 2333-2345. [Crossref]; d) Bavetsias V, Faisal A, Crumpler S, Brown N, Kosmopoulou M et al. (2013) Aurora isoform selectivity: design and synthesis of imidazo[4,5-b]pyridine derivatives as highly selective inhibitors of Aurora-A kinase in cells. J Med Chem 56: 9122-9135. [Crossref]; e) Sun J, Lv XH, Qiu HY, Wang YT, Du QR et al. (2013) Synthesis, biological evaluation and molecular docking studies of pyrazole derivatives coupling with a thiourea moiety as novel CDKs inhibitors. Eur J Med Chem 68: 1-9. [Crossref]
- a) Ahmad P, Woo H, Jun KY, Kadi AA, Abdel-Aziz HA et al. (2016) Design, synthesis, topoisomerase I & II inhibitory activity, antiproliferative activity, and structure-activity relationship study of pyrazoline derivatives: An ATP-competitive human topoisomerase IIα catalytic inhibitor. Bioorg Med Chem 24: 1898-1908. [Crossref]; b) Alam R, Wahi D, Singh R, Sinha D, Tandon V et al. (2016) Design, synthesis, cytotoxicity, HuTopoIIα inhibitory activity and molecular docking studies of pyrazole derivatives as potential anticancer agents. Bioorg Chem 69: 77-90. [Crossref]; c) H XianFeng Huang, L Xiang, Z Yong, S Guo-Qiang, H Qi-Long et al. (2012) Bioorg Med Chem 20: 4895-4900.
- MR Michaelides (2010) PCT Int Appl WO 2010065825.
- Srikrishna D, Godugu C, Dubey PK (2018) A Review on Pharmacological Properties of Coumarins. Mini-Rev Med Chem 18: 113-141. [Crossref]
- Kumar S, Bawa S, Gupta H (2009) Biological activities of quinoline derivatives. Mini-Rev Med Chem 9: 1648-1654. [Crossref]
- a) Domling A (2006) Recent developments in isocyanide based multicomponent reactions in applied chemistry. Chem Rev 106: 17-89. [Crossref]; b) L Banfi, R Riva (2005) Org React 65: 1-140; c) Ramon DJ, Yus M (2005) Asymmetric multicomponent reactions (AMCRs): the new frontier. Angew Chem Int Ed Engl 44: 1602-1634. [Crossref]; d) Zhu, J. Eur. J. Org. Chem. 2003, 1133-1144.
- a) RW Armstrong, AP Combs, PA Tempest, SD Brown, TA Keating (1996) Multiple-Component Condensation Strategies for Combinatorial Library Synthesis. Acc Chem Res 29: 123-131; b) Schreiber SL (2000) Target-oriented and diversity-oriented organic synthesis in drug discovery. Science 287: 1964-1969. [Crossref];
b) Magedov IV, Manpadi M, Slambrouck SV, Steelant WF, Rozhkova E et al. (2007) Discovery and Investigation of Antiproliferative and Apoptosis-Inducing Properties of New Heterocyclic Podophyllotoxin Analogues Accessible by a One-Step Multicomponent Synthesis. J Med Chem 50: 5183-5192. [Crossref]
c) S Werner, DM Turner, MA Lyon, DM Huryn, P Wipf (2006) A focused library of tetrahydropyrimidinone amides via a tandem Biginelli-Ugi multi-component process. Synlett: 2334-2338; d) B Nagaraju, J Kovvuri, KS Babu, VL Nayak, PR Adiyala et al. (2017) Tetrahedron 73: 6969-6976.
- a) Z Chen, J Bi, W Su (2013) Chin J Chem 31: 507-514; b) KV Sashidhara, GR Palnati, LR Singh, A Upadhyay, SR Avula et al. (2015) Green Chem. 17: 3766-3770.
- Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65: 55-63. [Crossref]
- Wu J, Zhao M, Qian K, Lee KH, Morris-Natschke S (2009) Novel N-(3-carboxyl-9-benzyl-beta-carboline-1-yl)ethylamino acids: synthesis, anti-tumor evaluation, intercalating determination, 3D QSAR analysis and docking investigation. Eur J Med Chem 44: 4153-4161. [Crossref]
- E Nyarko, N Hanada, A Habib, M Tabata (2004) Fluorescence and phosphorescence spectra of Au(III), Pt(II) and Pd(II) porphyrins with DNA at room temperature. Inorg Chim Acta 357: 739-745.
- Fukuda H, Katahira M, Tsuchiya N, Enokizono Y, Sugimura T et al. (2002) Unfolding of quadruplex structure in the G-rich strand of the minisatellite repeat by the binding protein UP1. Proc Natl Acad Sci U S A 99: 12685-12690. [Crossref]
- Shahabadi N, Kashanian S, Purfoulad M (2009) DNA interaction studies of a platinum(II) complex, PtCl(2)(NN) (NN=4,7-dimethyl-1,10-phenanthroline), using different instrumental methods. Spectrochim Acta A Mol Biomol Spectrosc 72: 757-761. [Crossref]
- JM Kelly, AB Tossi, DJ McConnell, C OhUigin (1985) A study of the interactions of some polypyridylruthenium (II) complexes with DNA using fluorescence spectroscopy, topoisomerisation and thermal denaturation. Nucleic Acids Res 13: 6017-6034. [Crossref]
- Metcalfe C, Rajput C, Thomas JA (2006) Studies on the interaction of extended terpyridyl and triazine metal complexes with DNA. J Inorg Biochem 100: 1314-1319. [Crossref]
- Ewesuedo RB, Ratain MJ (1997) Topoisomerase I Inhibitors. Oncologist 2: 359-364. [Crossref]
- Cao R, Peng W, Chen H, Ma Y, Liu X et al. (2005) DNA binding properties of 9-substituted harmine derivatives. Biochem Biophys Res Commun 338: 1557-1563. [Crossref]
- Schrö dinger suite 2014-3; Schrö dinger, LLC: New York, 2014.
- Staker BL, Feese MD, Cushman M, Pommier Y, Zembower D et al. (2005) Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J Med Chem 48: 2336-2345. [Crossref]
- Temperini C, Messori L, Orioli P, Di Bugno C, Animati F et al. (2003) The crystal structure of the complex between a disaccharide anthracycline and the DNA hexamer d(CGATCG) reveals two different binding sites involving two DNA duplexes. Nucleic Acids Res 31: 1464-1469. [Crossref]
- A Kamal, KS Babu, J Kovvuri, V Manasa, A Ravikumar, et al. (2015) Amberlite IR-120H: an efficient and recyclable heterogeneous catalyst for the synthesis of pyrrolo[1,2-a]quinoxalines and 5’H-spiro[indoline-3,4’ pyrrolo[1,2-a]quinoxalin]-2-ones. Tetrahedron Lett 56: 7012-7015.
- Botta M, Armaroli S, Castagnolo D, Fontana G, Pera P et al. (2007) Synthesis and biological evaluation of new taxoids derived from 2-deacetoxytaxinine J. Bioorg Med Chem Lett 17: 1579-1583. [Crossref]
- Tan C, Liu J, Chen L, Shi S, Ji L (2008) Synthesis, structural characteristics, DNA binding properties and cytotoxicity studies of a series of Ru (III) complexes. J Inorg Biochem 102: 1644-1653. [Crossref]
- Dexheimer TS, Pommier Y (2008) DNA cleavage assay for the identification of topoisomerase I inhibitors. Nat Protoc 3: 1736-1750. [Crossref]
