Anti-Microbial and Anti-Cancer Properties of Tat-IV13, A Hybrid Bi-Partite Peptide Containing The Short Non Active Iv13 Sequence of Human Ll37 Cathelecidin
A B S T R A C T
Therapeutic strategies based on optimization of the unique human LL37 cathelecin sequences including FK-16, the core active sequence of LL37, have already been proposed. In this study we have characterized Tat-IV13 a new host defense hybrid peptide, that combined YGRRKKRRQRRR, the hydrophobic N-terminal fragment of HIV-1 Tat47-57 cell penetrating sequence, with IV13, a short IVQRIKDFLRNLV inactive sequence resulting from the deletion of the three N-terminal amino acid residues of FK16. Tat- IV13 displayed potent host defense inhibitory effects leading both to the survival inhibition of U87G cells, a glioblastoma model, and to the inhibition of the growth of S. agalactiae NEM316 ?dltA strain, a Gram+ bacterial model. These results suggest that identification of hybrid specific Tat-cathelecidin peptides with high anti-tumor activity and anti-bactericidal activity may represent a powerful approach to identify new candidates for future therapeutic developments.
Keywords
Cationic sequences, cancer, bacteria
Introduction
Cationic anti-microbial peptides (CAMPs) are important components of the innate immune response with anti-infective and immunomodulatory activities [1, 2]. In this regard, LL-37 the active form of an unique human cathelecidin gene is a cationic, amphipathic peptide of 4.5 kDa with an a-helical structure that results from a proteinase 3 mediated proteolytic cleavage of a 18-kDa precursor HCT18 protein [3, 4]. HCAP-18 is mainly stored in neutrophil-speci?c granules, and LL37 can be detectable in body ?uids, including airway surface liquid, plasma, urine, breast milk and sweat [5]. LL-37 can exhibit a broad spectrum of antimicrobial activity against bacteria, fungi, and viral pathogens [3, 6]. Interestingly it has also been clearly established that both LL37 or its C-terminal fragment LL17-32, also termed FK16, exhibited cytotoxicity against distinct tumor cells [7-11]. In addition, we have recently reported that specific cellular or virally encoded sequences can display LL37-like host defense properties such as inhibition of U87G cells glioblastoma survival and inhibition of the growth of S. agalactiae NEM316 ?dltA strain, a Gram+ bacterial model [12, 13]. Furthermore, we also previously found that both FK16 alone or an hybrid bipartite peptide containing cell penetrating HIV-1 Tat47-57 and FK16 sequences display similar inhibitory effects against U87G cells glioblastoma and S. agalactiae NEM316 ?dltA strain [13].
In this study using structural modeling and functional studies we have characterized host defense properties of Tat-IV13 a new potential potent anti-tumor and anti-bacterial hybrid bi-partite peptide containing 24 amino acids (aa) residues combining the Tat penetrating and inactive IV13, a short inactive deleted FK16 sequence.
Materials & Methods
I Cell and peptides
We used human previously characterized Glioblastoma U87G [14] and NH2—biotinylated peptides (Proteogenix) prepared by solid-phase peptide synthesis, dissolved in DMSO and stored at -20C pending use.
II Sequence analyses and molecular modeling
Sequence alignment was based on FASTA programs and an algorithm based on helix-coil transition theory, AGADIR, was used to predict helical propensity [15] and previously used identify an amphipathic, helical region of LL-37 [16].
III Cytotoxicity assays
As previously described [14] a total of 3,000 cells were incubated for 24 hours with different concentrations of peptides and Cell cytotoxicity was analyzed by a colorimetric assay using 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (called MTT) for adherent cells as described by the manufacturer (Sigma).
IVBacterial Strains and antibacterial susceptibility test
S. Agalactiae mutant NEM 316 ?dltA strain, that is characterized by a complete absence of D-alanine due to the insertional inactivation of dltA, were previously described [17]. The Minimum Inhibitory Concentrations (MICs) of each peptide were tested in Todd- Hewitt broth (THB) buffered with 50 mM HEPES in 96-well Costar polypropylene microplates (Costar, Cambridge, USA) by a dilution method. Bacteria (10^6 CFU) were added in triplicates to wells containing increasing concentrations of the antimicrobial peptides. Plates were incubated 24h at 37°C and then read (OD600 nm) using microplate reader (Synergy 2, Biotek) for bacterial growth. The MICs90 was considered to be the peptide concentration that inhibited 90% growth.
Results
I Effect of FK16 cathelecidin mimetic cationic peptides on growth of S.Agalactiae NEM 316 ?dltA strain and on survival of U87G glioblastoma cells
We have first analyzed the effects of limited stepwise N-terminal deletions of FK16 on bacterial growth. As shown in Table 1, short N-terminal deletions of one (F16 in KR15) or two (F16+ K15 in RI14) residues reduced the wtFK16 inhibitory effect. In addition, deletion of the three FKR residues in IV13 suppressed the growth inhibition mediated by wtFK16. Furthermore, using MTT analyses, we have also monitored the effect of FK16 deletions on survival of U87G cells. As shown in Fig.1 upper panel, KR15, RI14 and 1V13 clearly counteracted the previously characterized strong inhibition of U87G survival mediated by wt FK16 [12,13].
Table 1: Acronym, sequences (N terminus to C terminus) and Effect of FK16 deletions on bacterial growth
|
Acronym |
Sequence |
Bacterial Inhibition*MIC90 |
|
FK16 |
FKRIVQRIKDFLRNLV |
6,25μM |
|
KR15 |
KRIVQRIKDFLRNLV |
25μM |
|
RI14 |
RIVQRIKDFLRNLV |
50μM |
|
IV13 |
IVQRIKDFLRNLV |
>100μM |
AA residues are expressed in one letter conventional code
*The MIC (μM) of each peptide is an average of triplicate measurements performed by a dilution method in 96-well polypropylene microplate. The MICs90 was considered to be the peptide concentration that inhibited growth of 90% of the tested strains.
Table 2: Acronym, net charge, predictive index of helicity (% AGADIR) and bacterial inhibition.
|
Acronym |
Net Charge |
Index of helicity (% AGADIR) |
Bacterial inhibition *MIC90 |
|
FK16 |
4 |
1,9 |
6,25μM |
|
Tat-FK16 |
10 |
1,85 |
6,25μM |
|
IV13 |
2 |
0,89 |
>100μM |
|
Tat-IV13 |
10 |
1,85 |
3,125μM |
|
Tat-rev IV13 |
10 |
1,43 |
6,25μM |
For Net Charge calculation see. An algorithm based on helix-coil transition theory, AGADIR, was used to predict helical propensity [16].
*The MIC (μM) of each peptide is an average of triplicate measurements performed by a dilution method in 96-well polypropylene microplate. The MICs90 was considered to be the peptide concentration that inhibited growth of 90% of the tested strains.
We hypothesized that the penetrating cell penetrating sequence of HIV1 Tat protein, the Tat47-57 sequence, combined with IV13 sequence may generate a biologically active bipartite peptide with FK16-like properties. To test this hypothesis, we synthesized hybrids Tat-IV13 and Tat-revIV13, the hybrid Tat peptide containing the reverse IV13 sequence initially designed as a potential negative control of the IV13 sequence. Furthermore, we comparatively analyzed the host defense properties of these peptides with FK16/Tat-FK16 (positive controls). Surprisingly, as shown in (Table 2) column 4, FK16, Tat-FK16 and Tat-revIV13 inhibited the growth of S.Agalactiae NEM 316 ?dltA strain with the same efficiency (MIC90=6,25?M), and surprisingly Tat-IV3 has a stronger anti-bacterial effect (MIC90=3,125?M). Interestingly, as shown in (Table 3), sequence alignment indicated that IV13 and revIV13 display a strong similarity (84,6%). And, consistently with the presence of a common Tat sequence, TAT-IV13 and TAT-revIV13 displayed a higher score (91,7%). Furthermore, structural bioinformatic analysis using the prediction AGADIR algorithm, suggested that FK16, Tat-FK16, Tat-IV3 and TatT-revIV3 are cationic peptide that displayed similar helical propensity (Table 2 column3) [15, 16]. Finally, functional MTT analyses illustrated in (Figure 1) lower panel indicated Tat-IV13 is much efficient than FK16 and Tat-revIV13 to inhibit U87G survival (estimated IC50= 3?M)
In conclusion these results indicated that Tat-revIV13 and FK16 displayed similar anti-bacterial effects MIIC90= 6,25?M). In addition, the results also suggested that TAT-IV13, used at a concentration of 3?M, is potential anti-tumor and anti-bacterial host defense peptide.
Table 3: Acronym, sequence alignment and similarity of IV3 mimetic peptides.
|
Acronym |
Sequence & alignment |
% similarity |
|
IV13
revIV13 |
IVQRIKDFLRNLV . . . : . : . : . . . VLNRLFDKIRQVI |
84,6 |
|
Tat-IV13
Tat-revIV13 |
YGRKKRRQRRRIVQRIKDFLRNLV : : : : : : : : : : : . . . : . : . : . . . YGRKKRRQRRRVLNRLFDKIRQVI |
91,7 |
AA residues are expressed in one letter conventional code. Sequence alignment and residues similarity were performed using FASTA [18].
Identification of Tat-IV13, a new potent host defense mimetic peptide derived from anti-microbial human FK16 cathelecidin sequence.
Figure 1: Effect of FK16 mimetic peptides on viability of U87G glioblastoma cells.
U87G cells were treated for 24 h with the different peptides and cell viability was assessed by MTT test (n=3). Upper panel: wildt type and N-Ter deleted FK16 sequences (0—25?M). Lower panel: hybrid cationic peptides containing Tat or FK16-derived sequences (0—25?M).
Discussion
Cathelicidin antimicrobial peptides such as human LL-37 and mouse mCRAMP are natural candidates antibiotics involved in innate immune defense [19]. Host defense strategies based on optimization of LL37 sequences have already been proposed [20]. In addition, identification of Tat peptides with high bactericidal activity is a promising therapeutical approach and recent studies highlighted the interests of TAT-modified cationic peptide for future development of novel antibiotics [21]. In this study using bioinformatics, including sequence alignment and modeling, and functional analyses we have characterized host defense properties of Tat-IV13 a new potential potent anti-tumor and anti-bacterial hybrid bi-partite peptide containing 24 amino acids (aa) residues combining Tat penetrating sequence with the short inactive FK16 deletion mutant named IV13. In addition, despite a high percentage of similarity with Tat-IV13, we also characterized Tat-revIV13 that displayed a similar and lower (MIC90=6,25?M). host defense properties with FK16 or Tat-FK16.
In conclusion, it is urgent to develop new antimicrobial strategies to counteract bacterial resistance to conventional antibiotics. In this regard we have identified TAT-IV13, a potential anti-tumor and anti-bacterial host defense peptide acting at micromolar concentrations (<3?M). Future work involving viruses, microbes and parasites, will be necessary to establish potential anti-infective effects of this molecule. In fine our data suggest that the design of specific Tat-cathelecidin hybrid peptides may be a useful strategy to generate new host defense molecules.
Acknowledgements
The present study was supported by Institute Pasteur
Conflict of interest Statement
The authors declare no conflict of interest.
Article Info
Article Type
Research ArticlePublication history
Received: Sun 08, Sep 2019Accepted: Thu 31, Oct 2019
Published: Thu 05, Dec 2019
Copyright
© 2023 Alphonse Garcia. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.CMR.2019.01.03
Author Info
Alphonse Garcia Bruno Périchon
Corresponding Author
Alphonse GarciaLaboratoire E3 des Phosphatases-Unité RMN
Figures & Tables
Table 1: Acronym, sequences (N terminus to C terminus) and Effect of FK16 deletions on bacterial growth
|
Acronym |
Sequence |
Bacterial Inhibition*MIC90 |
|
FK16 |
FKRIVQRIKDFLRNLV |
6,25μM |
|
KR15 |
KRIVQRIKDFLRNLV |
25μM |
|
RI14 |
RIVQRIKDFLRNLV |
50μM |
|
IV13 |
IVQRIKDFLRNLV |
>100μM |
AA residues are expressed in one letter conventional code
*The MIC (μM) of each peptide is an average of triplicate measurements performed by a dilution method in 96-well polypropylene microplate. The MICs90 was considered to be the peptide concentration that inhibited growth of 90% of the tested strains.
Table 2: Acronym, net charge, predictive index of helicity (% AGADIR) and bacterial inhibition.
|
Acronym |
Net Charge |
Index of helicity (% AGADIR) |
Bacterial inhibition *MIC90 |
|
FK16 |
4 |
1,9 |
6,25μM |
|
Tat-FK16 |
10 |
1,85 |
6,25μM |
|
IV13 |
2 |
0,89 |
>100μM |
|
Tat-IV13 |
10 |
1,85 |
3,125μM |
|
Tat-rev IV13 |
10 |
1,43 |
6,25μM |
For Net Charge calculation see. An algorithm based on helix-coil transition theory, AGADIR, was used to predict helical propensity [16].
*The MIC (μM) of each peptide is an average of triplicate measurements performed by a dilution method in 96-well polypropylene microplate. The MICs90 was considered to be the peptide concentration that inhibited growth of 90% of the tested strains.
Table 3: Acronym, sequence alignment and similarity of IV3 mimetic peptides.
|
Acronym |
Sequence & alignment |
% similarity |
|
IV13
revIV13 |
IVQRIKDFLRNLV . . . : . : . : . . . VLNRLFDKIRQVI |
84,6 |
|
Tat-IV13
Tat-revIV13 |
YGRKKRRQRRRIVQRIKDFLRNLV : : : : : : : : : : : . . . : . : . : . . . YGRKKRRQRRRVLNRLFDKIRQVI |
91,7 |
AA residues are expressed in one letter conventional code. Sequence alignment and residues similarity were performed using FASTA [18].
Identification of Tat-IV13, a new potent host defense mimetic peptide derived from anti-microbial human FK16 cathelecidin sequence.
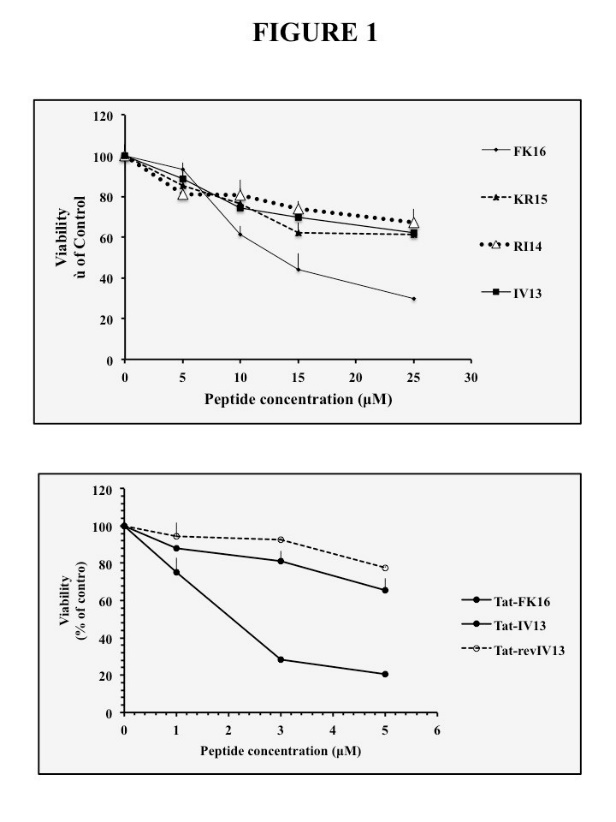
U87G cells were treated for 24 h with the different peptides and cell viability was assessed by MTT test (n=3). Upper panel: wildt type and N-Ter deleted FK16 sequences (0–25μM). Lower panel: hybrid cationic peptides containing Tat or FK16-derived sequences (0–25μM).
References
- Choi KY, Chow LN, Mookherjee N (2012) Cationic host defense peptides: multifaceted role in immune modulation and inflammation. J Innate Immun 4: 361-370. [Crossref]
- Bowdish DM, Davidson DJ, Hancock RE (2005) A re-evaluation of the role of host defense peptides in mammalian immunity. Curr Protein Pept Sci 6: 35-51. [Crossref]
- Sorensen O, Arnljots K, Cowland JB, Bainton DF, Borregaard N (1997) The human antibacterial cathelicidin, hCAP-18, is synthesized in myelocytes and metamyelocytes and localized to specific granules in neutrophils. Blood 90: 2796-2803. [Crossref]
- Sorensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS et al. (2001) Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3 Blood 97: 3951–3959. [Crossref]
- Bowdish DM, Davidson DJ, Hancock RE (2006) Immunomodulatory properties of defensins and cathelicidins. Curr Top Microbiol Immunol 306: 27-66. [Crossref]
- N.M. Frohm, B. Sandstedt, O.E. Sorensen, G. Weber, N. Borregaard, M. Stahle-Backdahl (1999) The human cationic antimicrobial protein (hCAP-18), a peptide antibiotic, is widely expressed in human squamous epithelia and colocalizes with interleukin-6. Infect Immun 67: 2561-2566. [Crossref]
- Wu WK, Wang G, Coffelt SB, Betancourt AM, Lee CW et al. (2010) Emerging roles of the host defense peptide LL-37 in human cancer and its potential therapeutic applications. Int J Cancer 127: 1741-1747. [Crossref]
- Mader JS, Mookherjee N, Hancock RE, Bleackley RC (2009) The human host defense peptide LL-37 induces apoptosis in a calpain- and apoptosis-inducing actor-dependent manner involving Bax activity. Mol Cancer Res 7: 689-702. [Crossref]
- 9. Wu WK, Sung JJ, To KF, Yu L, Li HT et al. (2010) The host defense peptide LL-37 activates the tumor-suppressing bone morphogenetic protein signaling via inhibition of proteasome in gastric cancer cells. J Cell Physiol 223: 178-186. [Crossref]
- Li X, Li Y, Han H, Miller DW, Wang G (2006) Solution structures of human LL-37 fragments and NMR-based identification of a minimal membrane targeting antimicrobial and anticancer region. J Am Chem Soc 128: 5776-5785. [Crossref]
- Ren SX, Cheng AS, To KF, Tong JH, Li MS et al. (2012) Host immune defense peptide LL-37 activates caspase-independent apoptosis and suppresses colon cancer. Cancer Res 72: 6512-6523. [Crossref]
- Colle JH, Périchon B, Garcia (2019) Antitumor and antibacterial properties of virally encoded cationic sequences. Biologics 13: 117-126. [Crossref]
- Périchon B, Garcia (2019) An Anti-infective properties of anti-cancer cationic peptides containing Survivin or Apolipoprotein E sequences. J Biotechnol Biomed 2: 161-168.
- Colle JH, Falanga PB, David-Watine B, Dutreix, M, Garcia A (2015) FTY720 overcomes resistance of human U87G Glioma cells expressing irradiation-induced SA-β-beta-gal biomarker. Current Topics in Pharmaclogy 19: 13-19.
- Lacroix E, A. R. Viguera, and L. Serrano (1998) Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic strength dependence and prediction of NMR parameters. J Mol Biol 284: 173-191. [Crossref]
- Sigurdardottir T, Andersson P, Davoudi M,Malmsten M, Schmidtchen A et al. (2006) In Silico Identification and Biological Evaluation of Antimicrobial Peptides Based on Human Cathelicidin LL-37. Antimicrob Agents Chemother 50: 2983-2989. [Crossref]
- Saar-Dover R, Bitler A, Nezer R, Shmuel-Galia L, Firon A et al. (2012) D-alanylation of lipoteichoic acids confers resistance to cationic peptides in group B streptococcus by increasing the cell wall density. PLoS Pathog 8: e1002891. [Crossref]
- Pearson WR (2014) BLAST and FASTA similarity searching for multiple sequence alignment. Methods Mol Biol 1079: 75-101. [Crossref]
- Zanetti, M. (2005) The role of cathelicidins in the innate host defenses of mammals. Curr Issues Mol Biol 7: 179-196. [Crossref]
- Fjell CD, Hiss JA, Hancock, RE, Schneider G (2012) Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov 11: 37-51. [Crossref]
- B He, Shiyi MA, Guifu Peng, Daohang He (2018) TAT-modified self-assembled cationic peptide nanoparticles as an efficient antibacterial agent. Nanomedecine 14: 365-372. [Crossref]
