Journals
Genomic Profiling for Patients with Solid Tumors: A Single-Institution Experience
A B S T R A C T
Background: Genomic tumor profiling is a novel technique that led to the identification of many genomic alterations in tumor tissues that could be exploited to deliver precise therapy to individual patient. Lack of data from Saudi Arabia about the utilization of that technology and its potential impact on clinical outcome has prompted this study.
Patients and Methods: Tumor tissues from 50 consecutive adult patients with metastatic solid cancer that is refractory to standard of care, were gnomically profiled.
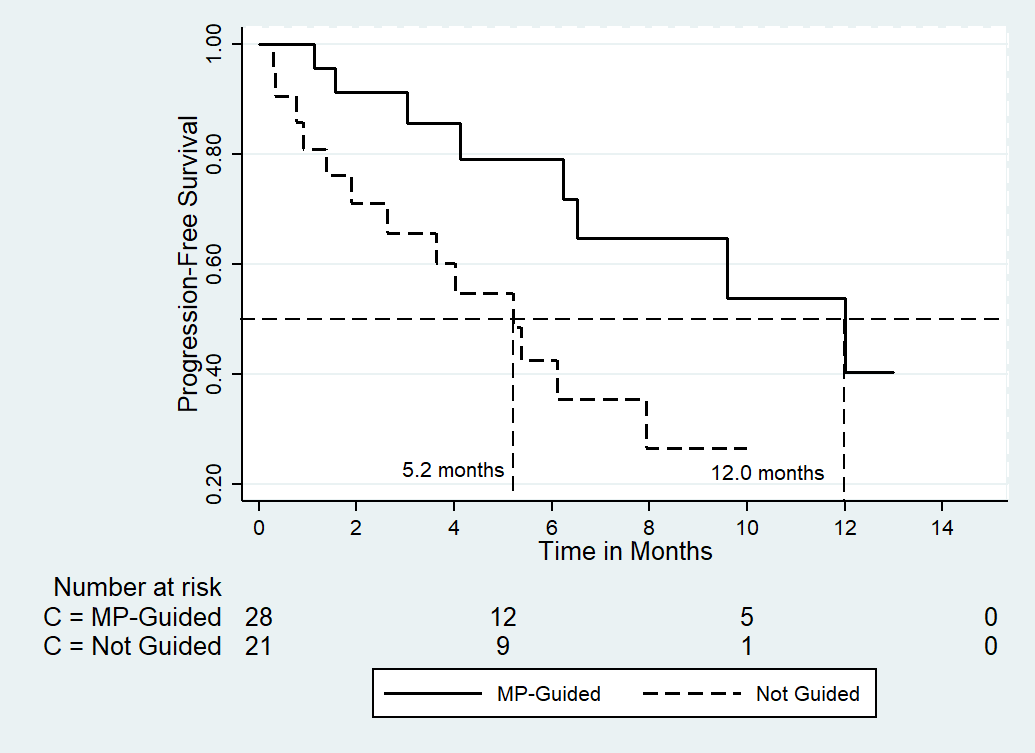
Results: Patients' median age was 56 years, and female constituted 76% of the series. All patients were heavily pretreated, with 52% having either breast, lung cancer, or ovarian cancer. In 88% of patients at least one genetic alteration was detected. Tumor profiling has guided the management decisions in 58%, 87%, and 14% of the overall patient population, breast cancer patients, and lung cancer patients, respectively. Meaningful disease response rates (complete remission, partial remission, and stable disease) were similar among those whose therapy decision was guided by tumor profiling (25 of 29 patients; 86%) and those where the therapy decision was not guided by the genomic findings (25 of 29 patients; 86% vs. 17 of 21 patients; 81%; P = 0.72). On the other hand, the median progression-free survival (PFS) determined from the time of making therapy decision based on the tumor profiling results was significantly longer among those whose management was supported by the findings (12.0 vs. 5.2 months, respectively; the hazard ratio and its 95% CI was 0.32 [0.13-0.81]; P = 0.017). While overall survival difference could not be estimated, the 12-months survival was 64% vs. 53% in the supported and the unsupported groups, respectively.
Conclusion: This preliminary experience demonstrated the feasibility and the clinical benefit of tumor profiling for cancer patients in Saudi Arabia. Tumor profiling is a promising novel technology; however, further research is required to address some of the inherent challenges to achieve the desired benefit.
Keywords
Cancer, neoplasm, genomic Profiling, tumor Profiling
Introduction
At the present era of precision medicine, recent advances in genomic technology have led to the identification of actionable or targetable genomic alterations in tumor tissues that could be exploited using specific therapy for each individual cancer patient [1]. Earlier data derived from phase I and retrospective studies demonstrated favorable outcome using such strategy [2, 3]. More recently, a prospective, single-center study conducted in patients with diverse refractory cancers who underwent comprehensive genomic profiling, the study group designed a calculating matching scores, based on the number of drug matches and genomic aberrations per patient [4]. The authors reported that patients with higher matching scores achieved higher response rate, and significantly longer progression-free survival (PFS) and overall survival (OS).
On the other hand, in the phase II SHIVA study, Le Tourneau et al. randomized 197 heavily pretreated patients with various solid tumors to receive a matched molecularly targeted agent or treatment at physician's choice [5]. The study could not demonstrate a PFS difference between the two groups. However, this study only included patients for whom a molecular alteration was identified within one of three molecular pathways (hormone receptor, PI3K/AKT/mTOR, and RAF/MEK). Furthermore, in a more recent update, the authors reported a 30% improvement in PFS in patients who crossed-over from the control to the experimental arm [5]. To the best of our knowledge, there has been no data about the use of tumor profiling among cancer patients in Saudi Arabia or in the nearby countries, moreover, there are no local institutional or national guidelines that could direct clinicians about how they could exploit that emerging technology to achieve the best outcome for cancer patients. The lack of such information has provided an impetus to report our early experience of genomic profiling of a series of 50 consecutive, pretreated cancer patients.
Patients and Methods
Between May 2017 and April 2019, consecutive adult patients with any kind of metastatic solid tumor diagnosed at a single institution were included in the current study. Patients must have demonstrated refractoriness to standard of care and needed to have an Eastern Cooperative Oncology Group performance status of 0 or 1. The study allowed testing for molecular profiling either from a tissue obtained from the primary tumor site or from a metastatic lesion. We intended to analyze the first 50 successfully profiled tumors. Tumor profiling of patients was performed on archival fixed formalin paraffin-embedded tissue using either of the two commercially available next generation sequencing (NGS) platforms, i.e. FoundationOne® (Foundation Medicine, Inc.) or OncoDEEP® (OncoDNA, Inc). The NGS mutational analysis using the FoundationOne® was based on a panel of 315 cancer-related genes, while OncoDEEP® platform is based on NGS of 75 cancer-related genes, besides several immunohistochemistry tests including protein phosphorylation [4, 7].
The findings of the genomic profile for each patient were discussed at a multidisciplinary tumor board to recommend further management. The decisions taken are classified as: 1. uphold the current treatment as guided by the profiling results; 2. uphold the current treatment not guided by the profiling results; 3. recommend changing/initiating a different treatment as guided by the results; 4. recommend changing/initiating a different treatment due to disease progression; or 5. recommend changing/initiating a different treatment as guided by the disease progression plus the results of genomic profiling. Either the first, the third, or the fifth decision was considered as a decision guided by the results of the genomic profiling. Otherwise, the decision was regarded as not supported by the tumor profile.
Response to therapy post tumor profiling was assed according to the RECIST (Response Evaluation Criteria In Solid Tumors) criteria and were reported as best response [8]. Comparing clinical benefit in disease response (complete response + partial response + stable disease) between management supported by tumor profiling vs. management not supported, was made using Fisher's exact test. PFS and OS were defined as interval between the date of implementing a therapeutic decision post tumor profiling to the date of progression, death, or date of last contact, as appropriate. PFS and OS curves were estimated using the Kaplan-Meier method, while the log-rank test was used to evaluate difference in survival between patients managed based on the tumor profiling results versus those whose management could not be guided by the genomic profiling. The Cox proportional hazard model was used to estimate hazard ratio (HR) and its 95% confidence interval (CI). All tests were two-sided at the 5% significance level. All statistical analyses were done with SPSS statistical package (IBM SPSS Statistics for Windows, version 25.0., New York, USA).
Table 1: Patients and disease characteristics
|
|
Number (%) |
|
Median age (95% CI), years |
56 (51.3-57.9) |
|
Gender Male Female |
12 (24) 38 (76) |
|
Diagnosis Breast Lung Ovary Colorectal Primary Unknown Pancreas Sarcoma Endometrium Others |
15 (30) 7 (14) 4 (8) 4 (8) 4 (8) 3 (6) 2 (4) 2 (4) 9 (18) |
|
Prior therapy Surgery Chemotherapy Endocrine therapy Her-2 targeted therapy CDK 4/6 inhibitors Checkpoint inhibitors Bevacizumab EGFR inhibitors Others |
31 (62) 36 (72) 14 (28) 4 (8) 11 (22) 7 (14) 6 (12) 4 (8) 5 (10) |
|
Current therapy when tumor profiling was performed Chemotherapy Endocrine therapy Immunotherapy HER-2 targeted therapy PARP inhibitor Anti-EGFR None |
17 (34) 7 (14) 4 (8) 3 (6) 1 (2) 1 (2) 17 (34) |
|
Median number of prior therapy lines (range) Chemotherapy Endocrine therapy |
2 (0 – 7) 0 (0 – 4) |
Table 2: Results of the tumor profiling
|
|
Number (%) |
|
Mean interval between diagnosis and tumor profiling testing (95% CI), months |
35.0 (23.0-47.1) |
|
Tumor profiling method OncoDEEP® FoundationOne® |
27 (54) 23 (46) |
|
PD-L1 expression Positive (10%, 30%, 60%, 60%) Low-expression Not done |
4 (8) 23 (46) 23 (46) |
|
CD-8 expression Positive Negative Not done |
16 (32) 10 (20) 24 (48) |
|
Microsatellite instability Negative Not done |
46 (92) 4 (8) |
|
Tumor mutation burden High (≥10 megabase) Low (<10 megabase) Negative |
5 (10) 17 (34) 24 (48) |
|
At least one alteration Next generation sequencing Positive Negative Immunohistochemistry Positive Negative/not done |
44 (88) 6 (12)
21 (42) 29 (58) |
|
Most common alterations detected (% of all 135 detected alterations) |
|
|
TP53 PI3KCA APC FGFR1 RAS/KRAS PTEN BRCA2 ESR1 MYC CCND1 CDKN2A CYP2D6 RB1 BRAF AKT |
21 (15.6) 10 (7.4) 7 (5.2) 7 (5.2) 6 (4.4) 5 (3.7) 4 (3) 4 (3) 4 (3) 3 (2.2) 3 (2.2) 3 (2.2) 3 (2.2) 2 (1.5) 2 (1.5) |
Results
Before reaching the targeted 50 patients with successful tumor profiling, the procedure has failed in four patients due to inadequate tissue sample. (Table 1) shows the patients and disease characteristics of the 50 patients whose tumors were successfully profiled. Patients' median age was 56 years, and female gender dominates the present series (76%). More than half of the included patients (52%), had breast cancer, lung cancer, or ovarian cancer. As shown in (Table 1), most patients were heavily pretreated. (Table 2) shows the results of the tumor profiling. Testing was performed almost 3 years from initial diagnosis (median, 35 months). Most patients had low expression of programmed-death legend-1 (PD-L1), and their tumors showed low mutational burden and low microsatellite instable (MSI) disease. In 88% of patients at least one genetic alteration was detected, and TP53 and PIK3CA were the most commonly identified aberrations (23%). Based on the results of the tumor profiling, multidisciplinary therapeutic decisions were made (Table 3). The table shows that tumor profiling has guided the management decisions in 58%, 87%, and 14% of the overall patient population, breast cancer patients, and lung cancer patients, respectively. (Table 3) also shows that the genomic profiling supported upholding patient's current treatment in 32% and 40%, for all patients and for breast cancer patients, respectively.
The median follow-up at the time of the analysis was 10.6 months estimated from the date of obtaining the genomic results and making therapeutic decision. As shown in (Table 4), there were no significant difference in the clinical benefit between those who had their management as supported by tumor profiling versus those who were treated in the absence of a such support (P = 0.72). On the other hand, as shown in (Fig. 1) and (Table 4), a significant PFS was demonstrated among patients who had their treatment has been guided by the tumor profiling with an absolute difference in median PFS of approximately 7 months (P = 0.01). The HR and its (95% CI) was 0.32 (0.13-0.81), indicating a 68% reduction in the risk of progression in favor of tumor profiling-based interventions. Like the advantage observed in the entire population, patients with metastatic breast cancer managed with the tumor profiling guidance attained a significant PFS benefit (Table 4).
At the time of the analysis, 7 of 29 (24%), and 7 of 21 (33%) patients were dead due to disease progression in the tumor profiling-guided and the non-guided groups, respectively. While the difference in OS was not calculable in both groups, the 1-year survival rates were 64% and 53%, respectively. To demonstrate the potential clinical benefit of tumor profiling if performed for an appropriate candidate, we briefly present two illustrative cases.
Case study 1
A 69-year old male was diagnosed with metastatic colon cancer on April 2017. His tumor was RAS wild-type. The patient received several lines of chemotherapy including FOLFOX, FOLFIRI, capecitabine, trifluridine and tipiracil, and regorafenib each in combination with anti- EGFR therapy. The tumor showed MSI- low, negative PD-L1 expression, low tumor mutational burden, and TP53, APC, and CDKN2A genomic alterations. CDKN2A gene is known as cyclin-dependent kinase (CDK) inhibitor 2A that codes for p16 protein to inhibit CDK 4/6. Therefore, having no other potentially effective therapy, we discussed with the patient the option of using palbociclib (anti CDK 4/6 inhibitor) as an off-label agent as suggested by the genomic results. The treatment was initiated after obtaining the patient's consent. The patient expressed subjective improvement, together with improvement in liver function tests and drop in the carcinoembryonic antigen. The benefit, however, only lasted for 6 weeks followed by disease progression.
Table 3: Clinical decision based on the results of tumor profiling
|
|
All (50 patients) |
Breast Cancer (15 patients) |
Lung Cancer (7 patients) |
|||
|
|
Guided by Tumor Profiling |
Not Guided by Tumor Profiling |
Guided by Tumor Profiling |
Not Guided by Tumor Profiling |
Guided by Tumor Profiling |
Not Guided by Tumor Profiling |
|
|
Patients Number (%) |
|||||
|
|
|
|
|
|
|
|
|
Uphold the current treatment guided by tumor profiling results |
16 (32) |
- |
6 (40) |
- |
1 (14) |
5 (72) |
|
|
|
|
|
|
|
|
|
Uphold the current treatment not guided by tumor profiling results |
- |
19 (38) |
- |
2 (13) |
- |
- |
|
|
|
|
|
|
|
|
|
Change or initiate treatment guided by tumor profiling results |
4 (8) |
- |
- |
- |
- |
- |
|
|
|
|
|
|
|
|
|
Change or initiate treatment due to disease progression |
- |
2 (4) |
- |
- |
- |
1 (14) |
|
|
|
|
|
|
|
|
|
Change or initiate treatment due to tumor profiling results plus disease progression |
9 (18) |
- |
7 (47) |
- |
- |
- |
|
|
|
|
|
|
|
|
|
Total |
29 (58) |
21 (42) |
13 (87) |
2 (13) |
1 (14) |
6 (86) |
Figure 1
Table 4: Analysis of the potential benefits for tumor profiling
|
|
Treatment Decision Guided by Tumor Profiling (29 Patients) |
Treatment Decision not Guided by Tumor Profiling (21 Patients) |
P value |
|
|
|
|
|
|
Initial response CR + PR + SD Progression |
25 (86) 4 (14) |
17 (81) 4 (19) |
0.72 |
|
|
|
|
|
|
Median progression-free survival (95% CI), months |
|
|
|
|
Overall |
7.9 (3.6-12.3) |
|
|
|
Tumor profiling effect |
12.0 (8.4-15.6) |
5.2 (2.9-7.4) |
0.01 |
|
Hazard ration of PFS (95% CI) |
0.32 (0.13-0.81) |
0.017 |
|
|
|
|
|
|
|
Median overall survival (95% CI), months |
|
|
|
|
Overall |
17.9 (5.4-30.4) |
|
|
|
Tumor profiling effect |
Not reached |
Not reached |
Not calculable |
|
|
|
|
|
|
12-month overall survival (SE) |
64% (13%) |
53% (11%) |
|
|
|
|
|
|
|
Breast cancer median progression-free survival (95% CI), months |
|
|
|
|
Overall |
12.0 (2.4-21.6) |
|
|
|
Tumor profiling effect |
12.0 (0.0-27.4) |
2.6 (not calculable) |
0.03 |
Case study 2
Tumor profiling identified BRCA2 mutation among three patients with breast cancer, in two patients that finding would have been missed as the two patients were not qualified for testing based on the National Comprehensive Cancer Network guidelines. A fourth patient was a female, aged 56 years. The patient had a breast cancer since the year 2008 and never experienced disease recurrence. Two years later she developed early endometrial cancer that was successfully managed. In the year 2017 she was found to have urinary bladder cancer with metastases to the lung. The patient developed disease progression on carboplatin-based therapy. A lung biopsy was done, and it was consistent with the urinary bladder primary. A tumor profiling done on that biopsy and it identified a BRCA2 mutation. The patient was started on olaparib - a PARB inhibitor, and she attained a durable remission that lasted for 13 months.
Discussion
In the current series of 50 consecutive pretreated patients with various solid tumors we demonstrated that tumor profiling has guided the management decisions in 58%, 87%, and 14% of the overall patient population, breast cancer patients, and lung cancer patients, respectively. The rate of guiding therapeutic decision based on tumor profiling was consistent with that reported in other series [2, 3, 9]. However, the percentage of our patients who were finally treated with a matched therapy was higher than that reported in other studies where less than 20% of patients actually received the matched therapy [9, 10]. The low tumor profiling guided-therapy in our lung cancer patients is consistent with the results reported by Jordan et al. where of 860 patients with metastatic lung adenocarcinoma, only 37% of patients received a matched therapy guided by their tumor molecular profile [11].
The illustrative example of the case study #1 showed that using an off-label of molecularly targeted agents may be useful although the benefit was short-lived. One may argue that have the patient had received palbociclib earlier and in combination with other approved agents, may be a longer response could have been achieved. On the other hand, based on the derived data from the SHIVA study the authors suggested that off-label use of agents based on tumor profiling should be discouraged [5]. Understandably, patients with different histology but having the same targetable aberration and exposed to the same agent may have different outcomes [12]. The current series showed that patients who received their treatment as guided by the tumor profiling findings showed an am improved PFS. More evident was the improvement in PFS among patients with breast cancer where there was an almost 10 months absolute difference in the median PFS in those who received their management according to their tumors' genomic profile versus those who had their therapy decision mainly based on physician's choice. Despite that demonstrated PFS benefit, tumor profiling-guided therapeutic decision was not associated with OS survival advantage. Unfortunately, failing to demonstrate an OS benefit using genomic profiling is not uncommon. In a large series that have used a unified platform (Caris Molecular Intelligence), patients whose treatments matched those predicted to be of benefit had an equal OS compared to those that did not [13].
Various series have reported diverse results concerning the impact of tumor profiling on decision-making and the also regarding the results of clinical benefit and the influence on patients' outcome [14]. There are several plausible explanations for such diversity such as the patient selection, naive patients versus those who were heavily pretreated, type of cancer, the time interval between initial diagnosis and when genomic profiling is performed, the platform used, archival versus fresh biopsy, the prevalence of targetable aberrations, and the proportion of patients including into clinical trials. The latter variable was addressed by the researchers at the Memorial Sloan Kettering Cancer Center where patients with diverse cancers were prospectively sequenced, and despite that 37% of tested tumors harboring a clinically relevant alteration, only 11% of more than 5,000 patients were enrolled onto genomically matched clinical trials [15]. In another prospective study of 250 patients with select solid tumors at the Cleveland Clinic, 49% of patients were recommended a specific therapy, but only 11% received such therapy [16].
Despite that the current report presents the only available data about implementing tumor profiling in cancer patients in Saudi Arabia, this series has some limitations. First, we have only included 50 patients with various tumor types. However, all studied patients had solid tumors and were all heavily pretreated. That diversity of patients' population has been the situation in many of the genomic profiling studies [2, 3, 5, 6, 13]. Moreover, the current series represents a preliminary analysis of a single-institutional experience to gain insight about that novel technology, its potential benefits, and its limitations. Second, in some of our patient we had to depend on archival tissue rather than on a fresh biopsy. The latter could have been more reliable as cancer molecular profile can change during disease evolution [17]. Nevertheless, while some genomic profiling studies mandated a fresh tumor biopsy, other studies relied on archival tissue, or allowed for archival or fresh-taken tissues [5, 3, 18].
In conclusion, precision medicine among cancer patients remains a major challenge for the oncology community but could enhance therapeutic options to be exploited in the advanced setting. Despite the potential benefit shown by us and by others, there is a long way to achieve the ideal desired effect. Future research should be able to address the most efficient and validated platform, the most reliable biomarkers that help selecting appropriate candidates, the most appropriate for when to perform genomic profiling technology throughout the course of the illness, exploiting the most reliable and validated fluid biopsy technique, and ways to lower the associated cost to make tumor profiling more cost-effective and affordable particularly for patients in developing and low-income countries [19, 20].
Article Info
Article Type
Research ArticlePublication history
Received: Thu 16, May 2019Accepted: Sun 23, Jun 2019
Published: Fri 16, Aug 2019
Copyright
© 2023 Ezzeldin M. Ibrahim. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.ACO.2019.02.04
Author Info
Ahmed A. Refae Ali M. Bayer Ezzeldin M. Ibrahim Ibrahim Mansoor Nasir A. Saleem Osama A. Al-Masri Rafat I. AbuShakra Wesal M. Eldahna
Corresponding Author
Ezzeldin M. IbrahimInternational Medical Center, Jeddah 21451, Kingdom of Saudi Arabia
Figures & Tables
Table 1: Patients and disease characteristics
|
|
Number (%) |
|
Median age (95% CI), years |
56 (51.3-57.9) |
|
Gender Male Female |
12 (24) 38 (76) |
|
Diagnosis Breast Lung Ovary Colorectal Primary Unknown Pancreas Sarcoma Endometrium Others |
15 (30) 7 (14) 4 (8) 4 (8) 4 (8) 3 (6) 2 (4) 2 (4) 9 (18) |
|
Prior therapy Surgery Chemotherapy Endocrine therapy Her-2 targeted therapy CDK 4/6 inhibitors Checkpoint inhibitors Bevacizumab EGFR inhibitors Others |
31 (62) 36 (72) 14 (28) 4 (8) 11 (22) 7 (14) 6 (12) 4 (8) 5 (10) |
|
Current therapy when tumor profiling was performed Chemotherapy Endocrine therapy Immunotherapy HER-2 targeted therapy PARP inhibitor Anti-EGFR None |
17 (34) 7 (14) 4 (8) 3 (6) 1 (2) 1 (2) 17 (34) |
|
Median number of prior therapy lines (range) Chemotherapy Endocrine therapy |
2 (0 – 7) 0 (0 – 4) |
Table 2: Results of the tumor profiling
|
|
Number (%) |
|
Mean interval between diagnosis and tumor profiling testing (95% CI), months |
35.0 (23.0-47.1) |
|
Tumor profiling method OncoDEEP® FoundationOne® |
27 (54) 23 (46) |
|
PD-L1 expression Positive (10%, 30%, 60%, 60%) Low-expression Not done |
4 (8) 23 (46) 23 (46) |
|
CD-8 expression Positive Negative Not done |
16 (32) 10 (20) 24 (48) |
|
Microsatellite instability Negative Not done |
46 (92) 4 (8) |
|
Tumor mutation burden High (≥10 megabase) Low (<10 megabase) Negative |
5 (10) 17 (34) 24 (48) |
|
At least one alteration Next generation sequencing Positive Negative Immunohistochemistry Positive Negative/not done |
44 (88) 6 (12)
21 (42) 29 (58) |
|
Most common alterations detected (% of all 135 detected alterations) |
|
|
TP53 PI3KCA APC FGFR1 RAS/KRAS PTEN BRCA2 ESR1 MYC CCND1 CDKN2A CYP2D6 RB1 BRAF AKT |
21 (15.6) 10 (7.4) 7 (5.2) 7 (5.2) 6 (4.4) 5 (3.7) 4 (3) 4 (3) 4 (3) 3 (2.2) 3 (2.2) 3 (2.2) 3 (2.2) 2 (1.5) 2 (1.5) |
Table 3: Clinical decision based on the results of tumor profiling
|
|
All (50 patients) |
Breast Cancer (15 patients) |
Lung Cancer (7 patients) |
|||
|
|
Guided by Tumor Profiling |
Not Guided by Tumor Profiling |
Guided by Tumor Profiling |
Not Guided by Tumor Profiling |
Guided by Tumor Profiling |
Not Guided by Tumor Profiling |
|
|
Patients Number (%) |
|||||
|
|
|
|
|
|
|
|
|
Uphold the current treatment guided by tumor profiling results |
16 (32) |
- |
6 (40) |
- |
1 (14) |
5 (72) |
|
|
|
|
|
|
|
|
|
Uphold the current treatment not guided by tumor profiling results |
- |
19 (38) |
- |
2 (13) |
- |
- |
|
|
|
|
|
|
|
|
|
Change or initiate treatment guided by tumor profiling results |
4 (8) |
- |
- |
- |
- |
- |
|
|
|
|
|
|
|
|
|
Change or initiate treatment due to disease progression |
- |
2 (4) |
- |
- |
- |
1 (14) |
|
|
|
|
|
|
|
|
|
Change or initiate treatment due to tumor profiling results plus disease progression |
9 (18) |
- |
7 (47) |
- |
- |
- |
|
|
|
|
|
|
|
|
|
Total |
29 (58) |
21 (42) |
13 (87) |
2 (13) |
1 (14) |
6 (86) |
Table 4: Analysis of the potential benefits for tumor profiling
|
|
Treatment Decision Guided by Tumor Profiling (29 Patients) |
Treatment Decision not Guided by Tumor Profiling (21 Patients) |
P value |
|
|
|
|
|
|
Initial response CR + PR + SD Progression |
25 (86) 4 (14) |
17 (81) 4 (19) |
0.72 |
|
|
|
|
|
|
Median progression-free survival (95% CI), months |
|
|
|
|
Overall |
7.9 (3.6-12.3) |
|
|
|
Tumor profiling effect |
12.0 (8.4-15.6) |
5.2 (2.9-7.4) |
0.01 |
|
Hazard ration of PFS (95% CI) |
0.32 (0.13-0.81) |
0.017 |
|
|
|
|
|
|
|
Median overall survival (95% CI), months |
|
|
|
|
Overall |
17.9 (5.4-30.4) |
|
|
|
Tumor profiling effect |
Not reached |
Not reached |
Not calculable |
|
|
|
|
|
|
12-month overall survival (SE) |
64% (13%) |
53% (11%) |
|
|
|
|
|
|
|
Breast cancer median progression-free survival (95% CI), months |
|
|
|
|
Overall |
12.0 (2.4-21.6) |
|
|
|
Tumor profiling effect |
12.0 (0.0-27.4) |
2.6 (not calculable) |
0.03 |
References
- Argani P, Perlman EJ, Breslow NE, Browning NG, Green DM et al. Servant N, Romejon J, Gestraud P, La Rosa P, Lucotte G et al. (2014) Bioinformatics for precision medicine in oncology: principles and application to the SHIVA clinical trial. Front Genet 5: 152. [Crossref]
- Von Hoff DD, Stephenson JJ Jr, Rosen P, Loesch DM, Borad MJ et al. (2010) Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin oncol 28: 4877-4483. [Crossref]
- Tsimberidou AM, Iskander NG, Hong DS, Wheler JJ, Falchook GS et al. (2012) Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res 18: 6373-6383. [Crossref]
- Wheler JJ, Janku F, Naing A, Li Y, Stephen B et al. (2016) Cancer Therapy Directed by Comprehensive Genomic Profiling: A Single Center Study. Cancer Res 76: 3690-3701. [Crossref]
- Le Tourneau C, Delord JP, Goncalves A, Gavoille C, Dubot C et al. (2015) Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 16: 1324-1334. [Crossref]
- Belin L, Kamal M, Mauborgne C, Plancher C, Mulot F et al. (2017) Randomized phase II trial comparing molecularly targeted therapy based on tumor molecular profiling versus conventional therapy in patients with refractory cancer: cross-over analysis from the SHIVA trial. Ann Oncol 28: 590-596. [Crossref]
- Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR et al. (2013) Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31: 1023-1031. [Crossref]
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D et al. (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45: 228-247. [Crossref]
- Massard C, Michiels S, Ferte C, Le Deley MC, Lacroix L et al. (2017) High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov 7: 586-595. [Crossref]
- Andre F, Bachelot T, Commo F, Campone M, Arnedos M et al. (2014) Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER). Lancet Oncol 15: 267-274. [Crossref]
- Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D et al. (2017) Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov 7: 596-609. [Crossref]
- Cecchini M, Walther Z, Sklar JL, Bindra RS, Petrylak DP et al. (2018) Yale Cancer Center Precision Medicine Tumor Board: two patients, one targeted therapy, different outcomes. Lancet Oncol 19: 23-24. [Crossref]
- Carter P, Alifrangis C, Cereser B, Chandrasinghe P, Belluz LDB et al. (2018) Does molecular profiling of tumors using the Caris molecular intelligence platform improve outcomes for cancer patients? Oncotarget 9: 9456-9467. [Crossref]
- Capdevila J, Rojo F, González-Martín A, Grande E, Martín-Algarra S et al. (2017) Molecular Profiling for Clinical Decision Making in Advanced Cancer: A Clinical Appraisal. J Cancer Research Treatment 5: 77-85.
- Zehir A, Benayed R, Shah RH, Syed A, Middha S et al. (2017) Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23: 703-713. [Crossref]
- Sohal DP, Rini BI, Khorana AA, Dreicer R, Abraham J et al. (2015) Prospective Clinical Study of Precision Oncology in Solid Tumors. J Natl Cancer Inst 108. [Crossref]
- Hedberg ML, Goh G, Chiosea SI, Bauman JE, Freilino ML et al. (2016) Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. J Clin Invest 126: 169-180. [Crossref]
- Middleton G, Crack LR, Popat S, Swanton C, Hollingsworth SJ et al. (2015) The National Lung Matrix Trial: translating the biology of stratification in advanced non-small-cell lung cancer. Ann Oncol 26: 2464-2469. [Crossref]
- Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P et al. (2018) Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 36: 1631-1641. [Crossref]
- Nelson B (2014) Genomic medicine: a question of value: despite the promise of personalized medicine, genomic testing has yet to prove its cost-effectiveness. Cancer Cytopathol 122: 557-558. [Crossref]
