Highlighting the Role of Cdk6 Associated MicroRNAs in Cancer Treatment Using In Silico Approaches
A B S T R A C T
MicroRNAs (miRNAs) are small non-coding RNA’s that controls the regulation of a gene. Due to the over expression or under expression of miRNAs it leads to cause tumor or any other type of cancers such as, melanoma, lymphoma, cardiovascular issue, breast cancer etc. So, miRNAs can be used as a drug target for cancer therapy. This study aimed to check binding cavities of microRNA's involved in regulation of CDK6 protein. There are 23 different families of miRNAs that are involved in regulation of CDK6. Each family has one or more miRNAs. All these miRNAs are involved in the up regulation or downregulation of a gene, which lead to different type of cancers. All miRNAs of each family docked with mRNA CDK6 protein. After performing in silico analysis of binding interactions of mRNA with miRNAs the results were further refined by their comparison with information regarding their energies, interaction of the mRNA and miRNAs. The results show that all miRNAs lie in Protein Kinase domain, but the residues that lie is different within the families and across the families.
Keywords
MicroRNA’s, CDK6, protein kinase domain, has-miR-1297
Introduction
MicroRNAs (miRNAs) are non-coding RNAs (ncRNAs) that control gene expression. miRNAs range from 17 to 24 nucleotide (nt) long RNAs. It has been known that miRNAs had ability to increase the expression of a target mRNA. Every single miRNA may target many different transcripts [1]. The alterations in miRNAs are associated with different human ailments, including cancer, immune disorders or cardiovascular issue. Their abnormalities are connected to start, leading towards metastases of human cancer. In human tumors it was found in no less than two autonomous reports distributed in the vicinity of 2004 and 2009 that 192 miRNAs were strangely communicated in tumor cells, including 168 overexpressed miRNAs, implying that high expression of miRNAs is a sign of harmful phenotype [2, 3]. As a treatment the principle RNA hindrance operators utilized till now in pre-clinical and clinical investigations include: antisense oligonucleotides (ASOs), ribozymes and the DNAzymes, small interfering RNAs (siRNAs) and short hairpin RNAs (shRNAs), and hostile to miRNA agents, for example, ASOs-against miRNAs, and Locked nucleic acids (LNA) against miRNAs sor antagomirs [2, 3].
The process to discover any drug is costly and lengthy. In the present era by the use of computers it becomes easier to improve the traditional strategies for RNA target lead identification [4-6]. There are different programs used which is called docking programs i.e. Auto Dock and Dock, it has been used for predicting the binding sites and affinities of ligands. Different RNA could be targeted with small molecules including ribosome, tRNA, mRNA, and so forth. The 3D structure of miRNA is essential for the in-silico studies. Once the miRNA structure is acquired, molecular docking-based virtual high-throughput screening (vHTS) strategies will be utilized to improve the miRNA drug discovery process in light of RNA-perfect scoring functions, in which it is important to re-assess the electrostatic association and solvation terms as per experimental analysis [7].
The aim of study was to perform in silico analysis of the binding interactions of mRNA and miRNAs is to know the binding sites of miRNA that binds with mRNA and with their ligands.
Materials and Methods
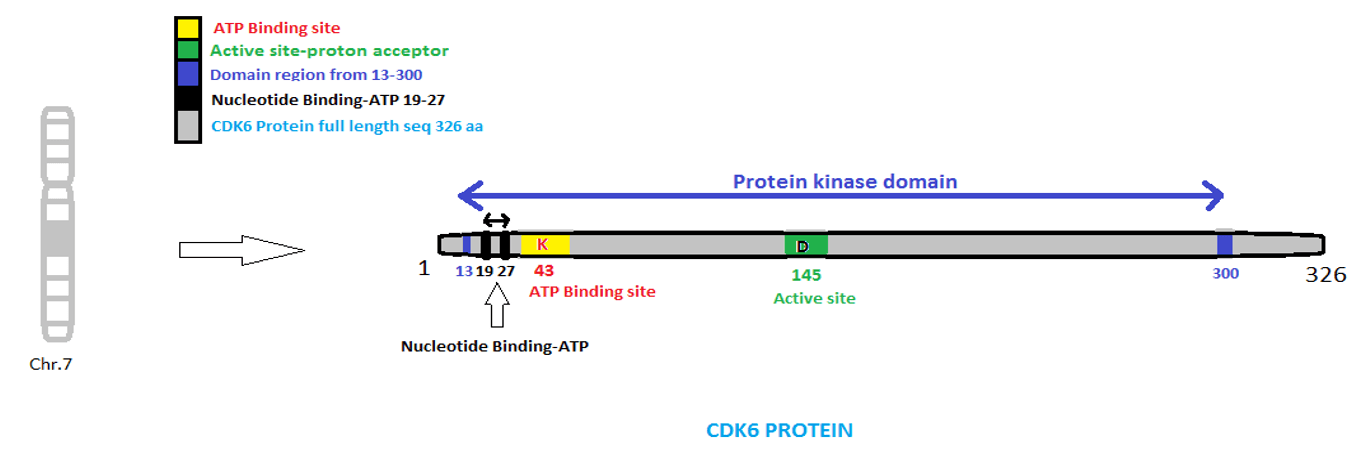
According to the literature there are many genes that are involved in the cancer. The genes that are involved in cancer causing in humans are mostly the Cell Dependent Kinases (CDKs) present in Cell Cycle. There are different genes that are present in CDKs at various phases. From the 38 genes checked in Diana Micro T web server, only CDK2, CDK4, CDK6, CDK9 and ATM have miRNAs that are associated with these genes, while rest of the genes does not find any miRNA that are associated with them. Among these genes CDK6 is the most significant gene that are involved in cancer. Cell Dependent Kinase 6 (CDK6) gene is present on the chromosome 7 in human. It has 326 amino acid residues which are encoded by 978 nucleotide bases. Table 1 shows the important regions of CDK6 while (Figure 1) graphically represent the features of CDK6 gene.
Table 1: Important features of CDK6 gene.
|
Feature keys |
Positions |
Descriptions |
|
Protein Kinase Domain |
13-300 |
CDK6 |
|
Binding site |
43 |
ATP |
|
Active site |
145 |
Proton Acceptor |
|
Nucleotide binding |
19-27 |
ATP |
|
Chain |
1-326 |
CDK6 |
Figure 1: Structure of CDK6 Gene.
I Ligand Dataset: miRNAs Identification
All miRNAs that are associated with CDK6 and involved in cancer causing to be searched by web server called Diana micro T version 5.0. There are 23 different miRNAs family that are involved in the regulation of CDK6 gene. These families have one or more than one miRNAs. All the miRNAs from each family are almost involved in the same pathways. The following (Table 2) shows miRNA’s families their members and the pathways in which miRNA’s are involved.
Table 2: miRNAs with their families and involved in different pathways.
|
Sr # |
Family |
Members(miRNAs) |
Pathways |
|
1 |
hsa-miR-1305/hsa-miR-1305 |
hsa-miR-1305 |
P53 signaling pathway Viral carcinogenesis Non-small cell lung Pathways in cancer Chronic myeloid Hepatitis B Melanoma Pancreatic Leukemia Cell cycle Measles Glioma Cancer |
|
2 |
hsa-miR-548c-5p/ miR-548-5p/559
|
hsa-miR-548a-5p/hsa-miR-548ab/vhsa-miR-548ad-5p/hsa-miR-548ae-5p/hsa-miR-548ak/hsa-miR-548am-5p/hsa-miR-548ap-5p/ hsa-miR-548aq-5p/ hsa-miR-548ar-5p/hsa-miR-548as-5p/hsa-miR-548au-5p/hsa-miR-548ay-5p/hsa-miR-548b-5p/hsa-miR-548bb-5p/ hsa-miR-548c-5p/hsa-miR-548d-5p/hsa-miR-548h-5p/hsa-miR-548i/hsa-miR-548j-5p/hsa-miR-548o-5p/hsa-miR-548w/hsa-miR-548y/hsa-miR-559 |
|
|
3 |
hsa-miR-4429 /miR-320/4429 |
hsa-miR-320a/hsa-miR-320b/has-miR-320c/hsa-miR-320d/hsa-miR-4429 |
|
|
4 |
hsa-miR-26b-5p/miR-26-5p/1297/4465 |
hsa_miR-26a-5p/hsa_miR-26b-5p/hsa_miR-1297/hsa_miR-4465 |
|
|
5 |
hsa-miR-548k/miR-548-5p/8054 |
hsa_miR-548av-5p/hsa_miR-548k/hsa_miR-8054 |
|
|
6 |
hsa-miR-107/miR103-3P/107 |
hsa-miR-103a-3p/hsa-miR-107 |
|
|
7 |
hsa-miR-21-3p/miR-21-3p/3591-p |
hsa-miR-21-3p/hsa-miR3-591-3p |
|
|
8 |
hsa-miR-590-3p/miR-590-3p |
has-miR-590-3p |
|
|
9 |
hsa-miR-362-3p/miR-329-3p/362-3p |
hsa-miR-329-3p/has-miR-362-3p |
|
|
10 |
hsa-miR-335-3p/miR-335-5p |
hsa-miR-335-3p |
|
|
11 |
hsa-miR-33a-5p/miR-33-5p |
hsa-miR33a-5p/hsa-miR33b-5p |
|
|
12 |
hsa-miR-3140-3p/miR-3140-3p |
hsa-miR-3140-3p |
|
|
13 |
hsa-miR-3619-5p/miR-214-3p/761/3619-5p |
hsa-miR-214-3p/has-miR-761/hsa-miR-3619-5p |
|
|
14 |
hsa-miR-32-3p/miR-32-3p |
hsa-miR-32-3p |
|
|
15 |
hsa-miR-548e-3p/miR-548ar-3p/548ar-3p/548az-3p/548e-3p/548f-3p |
hsa-miR-548a-3p/hsa-miR-548ar-3p/hsa-miR-548az-3p/hsa-miR-548e-3p/hsa-miR-548f-3p |
|
|
16 |
hsa-miR-576-5p/miR-576-5p |
hsa-miR-576-5p |
|
|
17 |
hsa-miR-548o-3p/miR-548-3p/1323 |
hsa-miR-548o-3p/hsa-miR-1323 |
|
|
18 |
hsa-miR-320e/miR-320 |
hsa-miR-320e |
|
|
19 |
hsa-miR-424-5p/miR-15-5p/16-5p/195-5p/424-5p/497-5p/6838-5p |
hsa-miR-15a-5p/hsa-miR-15b-5p/hsa-miR-16-5p/hsa-miR-195-5p/has-miR-424-5p/hsa-miR-497-5p/hsa-miR-6838-5p |
|
|
20 |
hsa-miR-450b-5p/miR-450-5p |
hsa-miR-450b-5p |
|
|
21 |
hsa-miR-142-5p/miR-142-5p/5590-3p |
hsa-miR-142-5p/hsa-miR-5590-3p |
|
|
22 |
hsa-miR-29b-3p/miR-29-3p |
hsa-miR-29a-3p/hsamiR-29b-3p/hsa-miR-29c-3p |
|
|
23 |
hsa-miR-589-3p/miR-589-3p |
hsa-miR-589-3p |
II miRNA 2D and 3D Structure Prediction
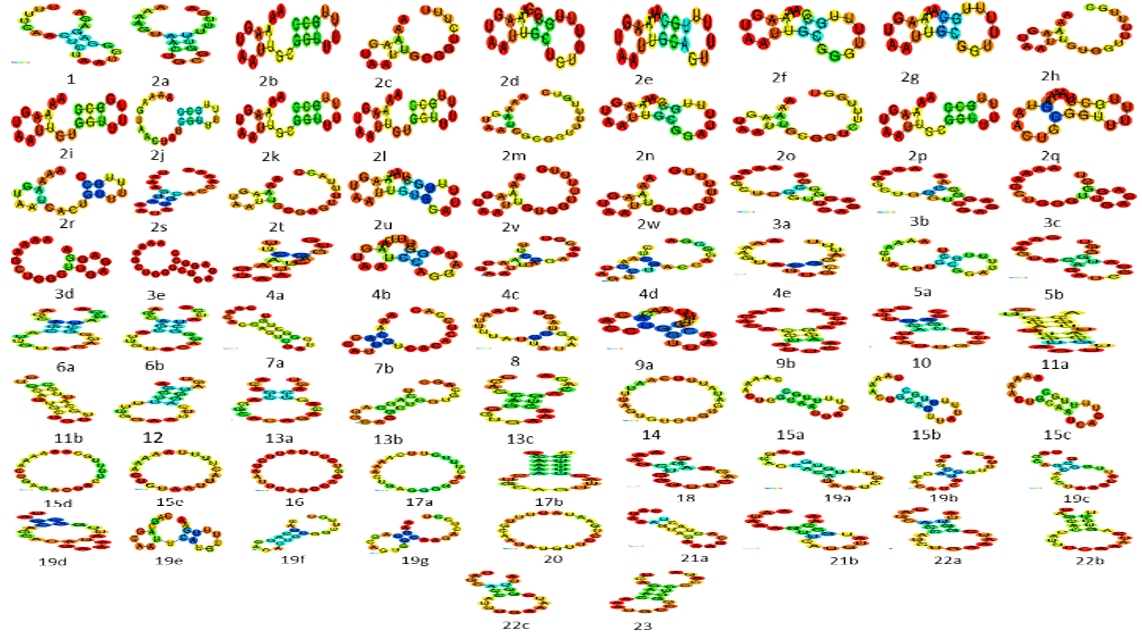
miRNAs 3D structure is predicted by using a web server called RNA Composer (Link) [8] . That server predicts the miRNA tertiary structure by giving input sequence of it. The sequence of miRNAs is present in a database called miRbase (Link) [9]. The tertiary structure of mRNA (CDK6) is also predicted by RNA Composer and using some more additional steps for its prediction, due to a large sequence. Centroid Fold is used for predicting secondary structure of RNA. It is used for predicting the secondary structure of all miRNAs. The following (Figure 2) shows the predicted secondary structure of miRNAs.
III Docking Studies
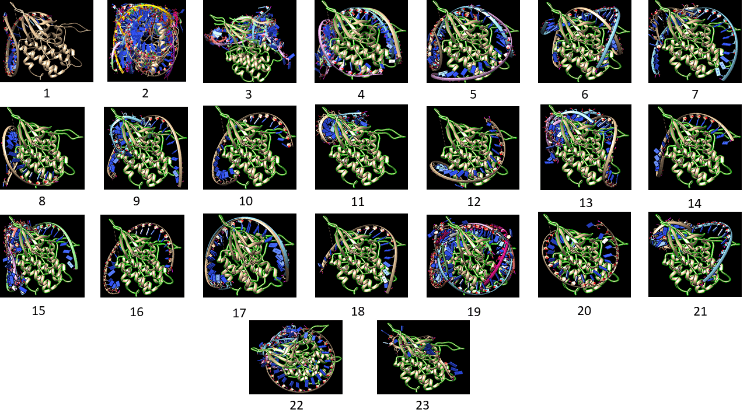
CDK6 was docked with all the miRNAs of each family for identifying the residues involved in hydrogen bonding and hydrophobic interaction within the families. For this purpose, Auto dock 4 software is used in which all the torsion angles in the small molecules were set free to perform flexible docking [10]. The grid was placed on CDK6 macromolecule that should cover the whole macromolecule and be large enough to allow the ligand to fully rotate and the empirical free energy function and the Lamarckian genetic algorithm (LGA) were used for docking where number of runs and population size were set to 50 [11]. Out of 23 families, family 4 has a miRNA which has the least binding energy and is considered as best conformation. This PDB file is viewed in LigPlot+ for observing the residues involved in Hydrogen bonding as well as in hydrophobic interactions [12]. Chimera view is used for interactive representation of docking results [13].
Results
I miRNA Secondary Structure Prediction
Secondary structure of all 76 miRNAs of all families are predicted by using a CentroidFold web server. miRNAs have flanking regions, loop, stems and bulges in their structures. The following table shows the important regions of miRNAs which has been observed after predicting their secondary structure. The (Table 3) below shows the important regions of miRNAs that is present in their structure.
Table 3: Different regions present in secondary structure.
|
Sr # |
Family |
Members(miRNAs) |
Flanking Regions |
Stems |
Loops |
Bulges |
Seed Region |
|
1 |
hsa-miR-1305/ hsa-miR-1305 |
hsa-miR-1305 |
1-7/20-22 |
8-10/17-19 |
11-16 |
- |
UUUCAAC
|
|
2 |
hsa-miR-548c-5p/ miR-548-5p/559
|
a) hsa-miR-548a-5p |
1-2/11-22 |
3-4/9-10 |
5-8 |
- |
AAAGUAA |
|
b) hsa-miR-548ab |
1-2/22 |
3-4/9-13/9-21 |
5-8/14-18 |
- |
|||
|
c) hsa-miR-548ad-5p |
1-2/11-20 |
3-4/9-10 |
5-8 |
- |
|||
|
d) hsa-miR-548ae-5p |
1-2/11-20 |
3-4/9-10 |
5-8 |
- |
|||
|
e) hsa-miR-548ak |
1-6/17-21 |
7-9/14-16 |
10-13 |
- |
|||
|
f) hsa-miR-548am-5p |
1-2 |
3-4/9-10/13-15/20-22 |
5-8/16-19 |
11-12 |
|||
|
g) hsa-miR-548ap-5p |
1-2/11-19 |
3-4/9-10 |
5-8 |
- |
|||
|
h) hsa-miR-548aq-5p |
1-2/22 |
3-4/9-12/20-21 |
5-8/13-19 |
- |
|||
|
i) hsa-miR-548ar-5p |
1-2 |
3-4/9-13/19-21 |
5-8/14-18 |
- |
|||
|
j) hsa-miR-548as-5p |
1-2/22 |
3-4/9-12/20-21 |
5-8/13-19 |
- |
|||
|
k) hsa-miR-548au-5p |
1-2 |
3-4/9-12/20-21 |
5-8/13-19 |
- |
|||
|
l) hsa-miR-548ay-5p |
1-2/11-21 |
3-4/9-10 |
5-8 |
- |
|||
|
m) hsa-miR-548b-5p |
1-2 |
3-4/9-10/13-15/20-22 |
5-8/16-19 |
11-12 |
|||
|
n) hsa-miR-548bb-5p |
1-12 |
13-15/20-22 |
16-19 |
- |
|||
|
o) hsa-miR-548c-5p |
1-2 |
3-4/9-10/13-15/20-24 |
5-8/16-19 |
11-12 |
|||
|
p) hsa-miR-548d-5p |
1-2 |
3-4/9-10/13-15/20-22 |
5-8/16-19 |
11-12 |
|||
|
q) hsa-miR-548h-5p |
1-3/10-22 |
4/9 |
5-8 |
- |
|||
|
r) hsa-miR-548i |
1-2/22 |
3-4/9-12/20-21 |
5-8/13-19 |
- |
|||
|
s) hsa-miR-548j-5p |
1-2/11-22 |
3-4/9-10 |
5-8 |
- |
|||
|
t) hsa-miR-548o-5p |
1-2 |
3-4/9-10/13-15/20-22 |
5-8/16-19 |
11-12 |
|||
|
u) hsa-miR-548w |
1-4/23 |
5/12-15/20-22 |
6-11/16-19 |
- |
|||
|
v) hsa-miR-548y |
1-3/22 |
4/9/14-15/20-21 |
5-8/16-19 |
10-13 |
|||
|
w) hsa-miR-559 |
1-4/15-21 |
5-7/12-14 |
8-11 |
- |
|||
|
3 |
hsa-miR-4429/miR-320/4429 |
hsa-miR-320a |
1-8/21-22 |
9/11-12/17-18/20 |
13-16 |
10/19 |
AAAGCUG
|
|
a) hsa-miR-320b |
1-8/21-22 |
9/11-12/17-18/20 |
13-16 |
10/19 |
|||
|
b) has-miR-320c |
1-10/19-20 |
11-12/17-18 |
13-16 |
- |
|||
|
c) hsa-miR-320d |
1-10/19 |
11-12/17-18 |
13-16 |
- |
|||
|
d) hsa-miR-4429 |
1-9/20 |
10-12/17-19 |
13-16 |
- |
|||
|
4 |
hsa-miR-26b-5p/miR-26-5p/1297/4465 |
a) hsa_miR-26a-5p |
1-4/21 |
5/10-12/19-20 |
6-9/13-18 |
- |
UCAAGUA
|
|
b) hsa_miR-26b-5p |
1-2/15-21 |
3-5/10-11/14 |
6-9 |
12-13 |
|||
|
c) hsa_miR-1297 |
1-2/15-17 |
3-5/10-11/14 |
6-9 |
12-13 |
|||
|
d) hsa_miR-4465 |
1-2/14-22 |
3/6/11/13 |
7-10 |
4-5/12 |
|||
|
5 |
hsa-miR-548k/miR-548-5p/8054 |
a) hsa_miR-548av-5p |
1-7/14-18 |
8/10-11/13 |
--- |
- |
AAAGUAC |
|
b) hsa_miR-548k |
1-10/22 |
11-12/20-21 |
13-19 |
- |
|||
|
c)hsa_miR-8054 |
1-7/19-21 |
8-9/17-18 |
10-16 |
- |
|||
|
6 |
hsa-miR-107/miR103-3P/107 |
a) hsa-miR-103a-3p |
1-3/20-23 |
4-6/17-19 |
7-16 |
- |
GCAGCAU |
|
b) hsa-miR-107 |
1-3/20-23 |
4-6/17-19 |
7-16 |
- |
|||
|
7 |
hsa-miR-21-3p/miR-21-3p/3591-p |
a) hsa-miR-21-3p |
1-6/21 |
7-11/16-20 |
12-15 |
- |
AACACCA |
|
b) has-miR-3591-3p |
1-312-22 |
4-5/10-11 |
6-9 |
- |
|||
|
8 |
hsa-miR-590-3p/miR-590-3p |
has-miR-590-3p |
1-9/18-21 |
10/17 |
11-16 |
- |
AAUUUUA |
|
9 |
hsa-miR-362-3p/miR-329-3p/362-3p |
a) hsa-miR-329-3p |
1-3/18-22 |
4/9-11/16-17 |
5-8/12-15 |
- |
ACACACC |
|
b) has-miR-362-3p |
1-6/18-22 |
7-9/15-17 |
10-14 |
- |
|||
|
10 |
hsa-miR-335-3p/miR-335-5p |
hsa-miR-335-3p |
1-4/21-22 |
5-7/18-20 |
8-17 |
- |
UUUUCAU |
|
11 |
hsa-miR-33a-5p/miR-33-5p
|
a) hsa-miR33a-5p |
1-2/21 |
3-5/7-9/14-16/18-20 |
10-13 |
6/17 |
UGCAUUG |
|
b) hsa-miR33b-5p |
1-2 |
3-5/7-9/14-16/18-20 |
10-13 |
6/17 |
|||
|
12 |
hsa-miR-3140-3p/miR-3140-3p |
hsa-miR-3140-3p |
1/20-22 |
2-5/16-19 |
6-15 |
- |
GCUUUUG |
|
13 |
hsa-miR-3619-5p/miR-214-3p/761/3619-5p |
a) hsa-miR-214-3p |
1-3/20-22 |
4-5/18-19 |
6-17 |
- |
CAGCAGG
|
|
b) has-miR-761 |
1-4/18-22 |
5-7/15-17 |
8-14 |
- |
|||
|
c) hsa-miR-3619-5p |
1/16-22 |
2-5/12-15 |
6-11 |
- |
|||
|
14 |
hsa-miR-32-3p/miR-32-3p |
hsa-miR-32-3p |
- |
- |
1-22 |
- |
AAUUUAG |
|
15 |
hsa-miR-548e-3p/miR-548ar-3p/548ar-3p/548az-3p/548e-3p/548f-3p |
a) hsa-miR-548a-3p |
1-8 |
9-12/19-22 |
13-18 |
- |
AAAACUG |
|
b) hsa-miR-548ar-3p |
1-7 |
8-11/18-21 |
12-17 |
- |
|||
|
c) hsa-miR-548az-3p |
1-7 |
8-11/18-21 |
12-17 |
- |
|||
|
d) hsa-miR-548e-3p |
- |
- |
1-22 |
- |
|||
|
e) hsa-miR-548f-3p |
- |
- |
1-19 |
- |
|||
|
16 |
hsa-miR-576-5p/miR-576-5p |
hsa-miR-576-5p |
- |
- |
1-22 |
- |
UUCUAAU |
|
17 |
hsa-miR-548o-3p/miR-548-3p/1323 |
a) hsa-miR-548o-3p |
1/22 |
2-6/17-21 |
7-16 |
|
CAAAACU |
|
b) hsa-miR-1323 |
- |
- |
1-22 |
- |
|||
|
18 |
hsa-miR-320e/miR-320 |
hsa-miR-320e |
1-4/18 |
5-6/16-17 |
7-15 |
- |
AAGCUGG |
|
19 |
hsa-miR-424-5p/miR-15-5p/16-5p/195-5p/424-5p/497-5p/6838-5p |
a) hsa-miR-15a-5p |
1-6 |
7-10/19-22 |
11-18 |
- |
AGCAGCA |
|
b) hsa-miR-15b-5p |
1-3/16-22 |
4-5/14-15 |
6-13 |
- |
|||
|
c) hsa-miR-16-5p |
1-5/22 |
6-7/20-21 |
8-19 |
- |
|||
|
d) hsa-miR-195-5p |
1-2 |
3-4/10/15/20-21 |
11-14 |
5-9/16-19 |
|||
|
e) has-miR-424-5p |
1-4/21-22 |
5/10/12-13/19-20 |
69/14-18 |
11 |
|||
|
f) hsa-miR-497-5p |
1-2/15-21 |
3-6/11-14 |
7-10 |
- |
|||
|
g) hsa-miR-6838-5p |
1-2/15-22 |
3-4/12-13 |
5-11 |
|
|||
|
20 |
hsa-miR-450b-5p/miR-450-5p |
hsa-miR-450b-5p |
- |
- |
- |
1-22 |
UUUGCAA
|
|
21 |
hsa-miR-142-5p/ miR-142-5p/5590-3p |
a) hsa-miR-142-5p |
1-5 |
6-10/17-21 |
11-16 |
- |
AUAAAGU
|
|
b) hsa-miR-5590-3p |
1-6/20-21 |
7-9/17-19 |
10-16 |
- |
|||
|
22 |
hsa-miR-29b-3p/miR-29-3p |
a) hsa-miR-29a-3p |
1-4/21-22 |
5-7/18-20 |
8-17 |
- |
AGCACCA
|
|
b) hsamiR-29b-3p |
1-2/22-23 |
3-6/19-21 |
7-18 |
- |
|||
|
c) hsa-miR-29c-3p |
1-4/21-22 |
5-7/18-20 |
18-17 |
- |
|||
|
23 |
hsa-miR-589-3p/miR-589-3p |
hsa-miR-589-3p |
1-3/20-24 |
4-7/16-19 |
8-15 |
- |
CAGAACA |
It is noticed that all the families detailed in (Table 3) have bulges, loops, Flanking regions and stems in their structure. The flanking regions are found at the start and end of the structures while the bulges, loops and stems are found in between. These secondary structures lie at different positions of the miRNA within and across the families.
Figure 2: Secondary structure of miRNAs.
II Docking Analysis of All Families
All members of each family have a sequence of not less than 17 nucleotides and not more than 24. The energy values differ within in the family and across the families as well, the energy values are not continuous. The protein residues present in all the families are not that much different. Most of the residues repeats in most of the miRNAs. The seed region, which is the important region of miRNAs, that interacts with mRNA is same within all the members of same family while it differs across the families. All the members of each family lie in the Protein Kinase (PK) domain, which ranges from 13-300. The flanking regions, stems and loops are present in almost all of the miRNAs. The flanking regions must lie in the start and end of the structure, stem, lop and bulges are almost present between the flanking regions.
III mRNA and miRNA Residues Mapping
The (Table 4) has the only miRNAs from each family having least binding energy that interact with CDK6 residues and their region.
Table 4: Mapping of CDK6 residues and miRNAs.
|
Sr # |
Family |
miRNA |
Energy |
Protein Residues Mapping + mRNA Mappiing |
miRNA Residues Mapping |
|
1 |
hsa-miR-1305/ hsa-miR-1305
|
hsa-miR-1305 |
-0.16 |
ARG90 = AGA + PK-Domain ARG87 = CGA + PK-Domain ARG78 = AGG + PK-Domain ARG186 = UAC + PK-Domain GLU72 = GAG + PK-Domain PHE135 = GAC + PK-Domain TYR292 = UCU + PK-Domain VAL181 = GUC + PK-Domain ARG144 = CAU + PK-Domain ALA167 = CUU + PK-Domain LEU166 = GGC + PK-Domain GLY165 = UUC + PK-Domain GLU21 = GAG + PK-Domain SER195 = UCC + PK-Domain SER194 = CAG + PK-Domain GLN193 = CUC + PK-Domain |
UUUUCAACU A=6,7 U=1,2,3,4 C=5,8 |
|
2 |
hsa-miR-548c-5p/ miR-548-5p/559 |
hsa-miR-548ap-5P |
-7.46 |
ARG46 = CGG + PK-Domain TYR24 = UAU + PK-Domain + NB-ATP LEU96 = UUA + PK-Domain GLN193 = CAG + PK-Domain SER195 = CUA + PK-Domain TYR196 = CGC + PK-Domain ARG144 = CGC + PK-Domain PRO244 = UAG + PK-Domain |
AAAAGUAAU A=2,3,4,7,8 G=5 U=6 |
|
3 |
hsa-miR-4429/miR-320/4429 |
hsa-miR-320b |
-1.2 |
VAL179 = GUG + PK-Domain ILE169 = AUC + PK-Domain SER171 = AGU + PK-Domain TYR170 = UAU + PK-Domain TRP184 = UGG + PK-Domain GLN103 = CAA + PK-Domain TYR185 = UAC + PK-Domain ARG168 = CGC + PK-Domain GLN149 = CAG + PK-Domain PRO17 = GCG + PK-Domain LYS216 = GCC + PK-Domain LEU152 = CUG + PK-Domain LEU166 = CUU + PK-Domain GLU18 = GAG + PK-Domain GLY20 = GGG + PK-Domain + NB-ATP ILE19 = AUC + PK-Domain + NB-ATP ALA17 = GCG + PK-Domain LYS111 = AAA + PK-Domain PHE164 = UUC + PK-Domain ASN150 = AAC + PK-Domain PRO148 = CCA + PK-Domain THR106 = ACC + PK-Domain ILE151 = AUU + PK-Domain ASP110 = GAU + PK-Domain HIS100 = CAU + PK-Domain ASP104 = GAC + PK-Domain LEU105 = UUG + PK-Domain VAL101 = GUC + PK-Domain ASP102 = GAU + PK-Domain |
AAAAGCUGGG A=2,3,4 G=5,8 C=6 U=7 |
|
4 |
hsa-miR-26b-5p/miR-26-5p/1297/4465
|
hsa-miR-1297 |
-10.63 |
GLU21 = GAG + PK-Domain + NB-ATP LEU166 = CUU + PK-Domain ALA167 = GCC + PK-Domain THR107 = ACU + PK-Domain ASP110 = GAU + PK-Domain LYS111 = AAA + PK-Domain TYR196 = CGC + PK-Domain ALA197 = CAC + PK-Domain ARG140 = CGA + PK-Domain THR198 = CCC + PK-Domain HIS137 = CAU + PK-Domain SER138 = UCA + PK-Domain |
UUCAAGUAA A=4,5,8 G=6 U=2,7 C=3 |
|
5 |
hsa-miR-548k/miR-548-5p/8054 |
hsa-miR-548av-5p |
-6.29 |
ALA167 = GCC + PK-Domain GLN193 = CAG + PK-Domain ILE169 = AUC + PK-Domain VAL181 = GUC + PK-Domain ARG186 = AGA + PK-Domain ARG78 = AGG + PK-Domain HIS139 = CAC + PK-Domain GLU72 = GAG + PK-Domain SER139 = CAC + PK-Domain PHE135 = UUU + PK-Domain TYR292 = CAG + PK-Domain |
AAAAGUACU A=2,3,4,7 G=5 U=6 C=8 |
|
6 |
hsa-miR-107/miR103-3P/107 |
hsa-miR-107 |
-1.2 |
VAL179 = GUG + PK-Domain GLN193 = CAG + PK-Domain SER155 = AGC + PK-Domain SER156 = AGC + PK-Domain TYR108 = UAC + PK-Domain LYS111 = AAA + PK-Domain VAL112 = GUU + PK-Domain THR121 = ACC + PK-Domain PRO113 = CCA + PK-Domain ARG140 = CGA + PK-Domain LEU94 = CUA + PK-Domain THR84 = ACA + PK-Domain THR92 = ACC + PK-Domain VAL47 = GUG + PK-Domain PRO118 = CCC + PK-Domain |
AGCAGCAUU A=4,7 G=2,5 U=8 C=3,6
|
|
7 |
hsa-miR-21-3p/miR-21-3p/3591-p
|
hsa-miR-3591-3p |
-5.8 |
GLN193 = CAG + PK-Domain SER194 = UAG + PK-Domain SER195 = CUA + PK-Domain ARG144 = CGC + PK-Domain ALA197 = CAC + PK-Domain TYR292 = CAG + PK-Domain |
AAACACCAU A=2,3,5,8 G= -- U= -- C=4,6,7 |
|
8 |
hsa-miR-590-3p/miR-590-3p
|
hsa-miR-590-3p |
-0.93 |
ARG140 = CGA + PK-Domain SER195 = CUA + PK-Domain ALA197 = CAC + PK-Domain ASN284 = CCC + PK-Domain ALA280 = UUU + PK-Domain ARG78 = AGG + PK-Domain GLY157 = GGA + PK-Domain SER156 = AGC + PK-Domain THR121 = ACC + PK-Domain PRO118 = CCC + PK-Domain GLU120 = GAA + PK-Domain |
UAAUUUUAU A=2,3,8 G= U=4,5,6,7 C=
|
|
9 |
hsa-miR-362-3p/miR-329-3p/362-3p
|
hsa-miR-329-3P |
-2.64 |
GLU21 = GAG + PK-Domain +NB-ATP LEU166 = CUU + PK-Domain ALA167 = GCC + PK-Domain LYS111 = AAA + PK-Domain PRO113 = CCA + PK-Domain GLN193 = CAG + PK-Domain ARG144 = CGC + PK-Domain SER195 = CUA + PK-Domain VAL142 = GUG + PK-Domain TYR196 = CGC + PK-Domain ALA197 = CAC + PK-Domain THR198 = CCC + PK-Domain HIS137 = CAU + PK-Domain SER138 = UCA + PK-Domain ARG140 = CGA + PK-Domain |
AACACACCU A=2,4,6, G= U= C=3,5,7,8 |
|
10 |
hsa-miR-335-3p/miR-335-5p
|
hsa-miR-335-3p |
0.12 |
SER194 = UAG + PK-Domain ASP246 = UGU + PK-Domain GLN193 = CAG + PK-Domain VAL179 = GUG + PK-Domain ILE169 = AUC + PK-Domain TYR292 = CAG + PK-Domain |
UUUUUCAUU A=7 G= U=2,3,4,5,8 C=6 |
|
11 |
hsa-miR-33a-5p/miR-33-5p
|
hsa-miR33a-5p |
-0.81 |
ASP246 = UGU + PK-Domain GLN193 = CAG + PK-Domain SER194 = UAG + PK-Domain SER195 = CUA + PK-Domain VAL190 = GUC + PK-Domain ARG144 = CGC + PK-Domain VAL181 = GUC + PK-Domain GLU189 = GAA + PK-Domain ALA197 = CAC + PK-Domain ALA23 = GCC + PK-Domain + NB-ATP GLY22 = GGC + PK-Domain + NB-ATP ARG140 = CGA + PK-Domain VAL142 = GUG + PK-Domain VAL141 = GUA + PK-Domain HIS143 = CAU + PK-Domain |
GUGCAUUGUAGUUGCAUUGCA A=5,16 G=3,8,14,19 U=2,6,7,13,17,18 C=4,15
|
|
12 |
hsa-miR-3140-3p/miR-3140-3p
|
hsa-miR-3140-3p |
-6.79 |
VAL16 = GUG + PK-Domain VAL17 = GCG + PK-Domain PRO74 = CCC + PK-Domain TYR292 = CAG + PK-Domain SER155 = AGC + PK-Domain SER156 = AGC + PK-Domain SER293 = UGC + PK-Domain LYS279 = GUG + PK-Domain SER290 = UGC + PK-Domain LYS111 = AAA + PK-Domain ALA286 = CAA + PK-Domain LYS287 = AAG + PK-Domain |
AGCUUUUGG A= G=2,8 U=4,5,6,7 C=3
|
|
13 |
hsa-miR-3619-5p/miR-214-3p/761/3619-5p
|
hsa-miR-214-3p |
-2.26 |
THR84 = ACA + PK-Domain LEU94 = CUA + PK-Domain VAL82 = GUG + PK-Domain ALA23 = GCC + PK-Domain + NB-ATP TYR24 = UAU + PK-Domain + NB-ATP GLY22 = GGC + PK-Domain + NB-ATP ARG140 = CGA + PK-Domain VAL142 = GUG + PK-Domain VAL141 = GUA + PK-Domain ARG144 = CGC + PK-Domain TYR196 = CGC + PK-Domain ARG186 = AGA + PK-Domain VAL181 = GUC + PK-Domain ILE169 = AUC + PK-Domain LEU166 = CUU + PK-Domain ALA167 = GCC + PK-Domain LYS111 = AAA + PK-Domain TYR108 = UAC + PK-Domain VAL112 = GUU + PK-Domain GLY116 = GGA + PK-Domain PRO113 = CCA + PK-Domain PRO115 = CCU + PK-Domain PRO118 = CCC + PK-Domain VAL117 = GUG + PK-Domain THR119 = ACU + PK-Domain |
ACAGCAGGC A=3,6 G=4,7,8 U= C=2,5 |
|
14 |
hsa-miR-32-3p/miR-32-3p
|
hsa-miR-32-3p |
-5.18 |
SER194 = UAG + PK-Domain ASP246 = CUA + PK-Domain GLN193 = CAG + PK-Domain LEU166 = CUU + PK-Domain ALA167 = GCC + PK-Domain ILE169 = AUC + PK-Domain ARG168 = CGC + PK-Domain TYR292 = CAG + PK-Domain |
CAAUUUAGU A=2,3,7 G=8 U=4,5,6 C= |
|
15 |
hsa-miR-548e-3p/miR-548ar-3p/548ar-3p/548az-3p/548e-3p/548f-3p
|
hsa-miR-548e-3p |
-6.04 |
GLN193 = CAG + PK-Domain SER194 = UAG + PK-Domain SER195 = CUA + PK-Domain TYR196 = CGC + PK-Domain ALA197 = CAC + PK-Domain THR198 = CCC + PK-Domain ASP110 = GAU + PK-Domain LYS111 = AAA + PK-Domain |
AAAAACUGA A=2,3,4,5 G=8 U=7 C=6 |
|
16 |
hsa-miR-576-5p/ miR-576-5p
|
hsa-miR-576-5p |
-0.47 |
SER19 = AUC + PK-Domain TYR196 = CGC + PK-Domain ALA197 = CAC + PK-Domain ILE169 = AUC + PK-Domain |
AUUCUAAUU A=6,7 G= U=2,3,5,8 C=4 |
|
17 |
hsa-miR-548o-3p/miR-548-3p/1323 |
hsa-miR-1323 |
0.16 |
ARG140 = CGA + PK-Domain TYR196 = CGC + PK-Domain VAL142 = GUG + PK-Domain HIS137 = CAU + PK-Domain ALA23 = GCC + PK-Domain + NB-ATP GLU21 = GAG + PK-Domain + NB-ATP LEU166 = CUU + PK-Domain GLY165 = GGC + PK-Domain ARG168 = CGC + PK-Domain GLN149 = CAG + PK-Domain ASP110 = GAU + PK-Domain LYS216 = GCC + PK-Domain ARG215 = AAA + PK-Domain GLU114 = GAG + PK-Domain THR267 = AGA + PK-Domain |
CCAAAACUG A=3,4,5,6 G= U=8 C=2,7 |
|
18 |
hsa-miR-320e/miR-320
|
hsa-miR-320e |
-9.15 |
LEU166 = CUU + PK-Domain ALA167 = GCC + PK-Domain ILE169 = AUC + PK-Domain LYS111 = AAA + PK-Domain PRO113 = CCA + PK-Domain PRO118 = CCC + PK-Domain ALA23 = GCC + PK-Domain + NB-ATP ARG144 = CGC + PK-Domain SER195 = CUA + PK-Domain VAL142 = GUG + PK-Domain TYR196 = CGC + PK-Domain THR198 = CCC + PK-Domain ARG140 = CGA + PK-Domain SER138 = UCA + PK-Domain |
AAAGCUGGG A=2,3 G=4,7,8 U=6 C=5 |
|
19 |
hsa-miR-424-5p/miR-15-5p/16-5p/195-5p/424-5p/497-5p/6838-5p |
hsa-miR-15b-5p |
-5.65 |
VAL16 = GUG + PK-Domain TYR24 = UAU + PK-Domain + NB-TP ARG44 = CGC + PK-Domain ARG186 = AGA + PK-Domain ASP145 = GAU + PK-Domain HIS143 = CAU + PK-Domain ARG78 = AGG + PK-Domain GLU72 = GAG + PK-Domain LYS111 = AAA + PK-Domain SER155 = AGC + PK-Domain |
UAGCAGCAC A=2,5,8 G=3,6 U= C=4,7 |
|
20 |
hsa-miR-450b-5p/miR-450-5p |
hsa-miR-450b-5p |
-1.22 |
ARG46 = CGG + PK-Domain VAL47 = GUG + PK-Domain GLN48 = CAG + PK-Domain ALA23 = GCC + PK-Domain LEU94 = CUA + PK-Domain TYR24 = UAU + PK-Domain + NB-ATP VAL82 = GUG + PK-Domain LEU96 = UUA + PK-Domain ARG144 = CGC + PK-Domain ARG186 = AGA + PK-Domain ASP145 = GAU + PK-Domain-Active Site LEU79 = UUG + PK-Domain ARG78 = AGG + PK-Domain GLU72 = GAG + PK-Domain VAL16 = GUG + PK-Domain LYS111 = AAA + PK-Domain THR154 = ACC + PK-Domain GLN18 = GAG + PK-Domain SER155 = AGC + PK-Domain GLY157 = GGA + PK-Domain SER156 = AGC + PK-Domain |
UUUUGCAAU A=7,8 G=5 U=2,3,4 C=6
|
|
21 |
hsa-miR-142-5p/ miR-142-5p/5590-3p |
hsa-miR-142-5p |
-3.36 |
VAL147 = AAA + PK-Domain GLN48 = CAG + PK-Domain VAL45 = GUG + PK-Domain LEU94 = CUA + PK-Domain TYR24 = UAU + PK-Domain +NB-ATP ASP246 = UGU + PK-Domain GLN193 = CAG + PK-Domain ALA248 = CCU + PK-Domain VAL82 = GUG + PK-Domain LEU96 = UUA + PK-Domain LYS43 = AAG + PK-Domain + ATP-Binding Site ASP163 = GAC + PK-Domain VAL247 = UGC + PK-Domain SER194 = CCU + PK-Domain LEU79 = UUG + PK-Domain VAL142 = GUG + PK-Domain ALA162 = GCU + PK-Domain ARG140 = CGA + PK-Domain HIS143 = CAU + PK-Domain LEU161 = CUC + PK-Domain VAL141 = GUA + PK-Domain LEU136 = CUU + PK-Domain VAL76 = GUG + PK-Domain |
CAUAAAGUA A=2,4,5,6 G=7 U=3,8 C=
|
|
22 |
hsa-miR-29b-3p/miR-29-3p |
hsa-miR-29c-3p |
-6.13 |
VAL47 = GUG + PK-Domain LEU94 = CUA + PK-Domain ALA23 = GCC + PK-Domain + NB-ATP GLU21 = GAG + PK-Domain + NB-ATP GLY22 = GGC + PK-Domain + NB-ATP GLY165 = GGC + PK-Domain VAL181 = GUC + PK-Domain SER223 = AGA + PK-Domain ASP224 = UGU + PK-Domain ARG144 = CGC + PK-Domain ARG186 = AGA + PK-Domain ASP145 = GAU + PK-Domain LYS147 = AAA + PK-Domain VAL82 = GUG + PK-Domain ARG140 = CGA + PK-Domain ASP242 = CUG + PK-Domain THR198 = CCC + PK-Domain PRO199 = CGU + PK-Domain PHE283 = UAA + PK-Domain PRO238 = AGG + PK-Domain ARG288 = AAU + PK-Domain THR282 = AUU + PK-Domain ASN284 = CCC + PK-Domain PRO285 = AGC + PK-Domain ALA286 = CAA + PK-Domain LYS287 = AAG + PK-Domain ILE289 = AUC + PK-Domain LEU237 = CCC + PK-Domain GLY236 = ACU + PK-Domain LYS279 = GUG + PK-Domain |
UAGCACCAU A=2,5,8 G=3 U= C=4,6,7 |
|
23 |
hsa-miR-589-3p/miR-589-3p |
hsa-miR-589-3p |
8.2 |
LEU94 = CUA + PK-Domain CYS83 = UGC + PK-Domain VAL82 = GUG + PK-Domain THR95 = ACU + PK-Domain ASP81 = GAU + PK-Domain LEU96 = UUA + PK-Domain VAL97 = GUG + PK-Domain LEU79 = UUG + PK-Domain GLN103 = CAA + PK-Domain VAL77 = GUC + PK-Domain ALA41 = GCG + PK-Domain PHE98 = UUU + PK-Domain PHE28 = UUC + PK-Domain LYS43 = AAG + PK-Domain + ATP-Binding Site LEU42 = UUG + PK-Domain ALA23 = GCC + PK-Domain + NB-ATP LYS26 = AAG + PK-Domain + NB-ATP THR106 = ACC + PK-Domain + NB-ATP GLU21 = GAG + PK-Domain + NB-ATP GLU18 = GAG + PK-Domain VAL27 = GUG + PK-Domain + NB-ATP ALA17 = GCG + PK-Domain GLY20 = GGG + PK-Domain + NB-ATP VAL142 = GUG + PK-Domain ILE19 = AUC + PK-Domain + NB-ATP PHE164 = UUC + PK-Domain THR107 = ACU + PK-Domain HIS143 = CAU + PK-Domain LYS29 = AAG + PK-Domain VAL77 = GUC + PK-Domain HIS100 = CAU + PK-Domain GLU99 = GAA + PK-Domain ASP102 = GAU + PK-Domain ASP145 = GAU + PK-Domain ASN150 = AAC + PK-Domain LYS111 = AAA + PK-Domain ARG78 = AGG + PK-Domain ARG144 = CGC + PK-Domain ARG186 = AGA + PK-Domain ALA167 = GCC + PK-Domain VAL181 = GUC + PK-Domain VAL190 = GUC + PK-Domain THR182 = GAU + PK-Domain LEU146 = CUA + PK-Domain ARG168 = CGC + PK-Domain LYS147 = AAA + PK-Domain ILE169 = AUC + PK-Domain TYR185 = UAC + PK-Domain GLN149 = CAG + PK-Domain PRO148 = CCA + PK-Domain SER204 = GAG + PK-Domain ASP104 = GAC + PK-Domain GLN193 = CAG + PK-Domain |
UCAGAACAA A=3,5,6,8 G=4 U= C=2,7
|
Figure 3: Shows interaction between mRNA (CDK6) with miRNAs of each family.
IV Observed Significant Features of mRNA and miRNAs after Docking
After the analysis of all the above results of mRNA and miRNAs all the complexes have different energy values, structures and their interacting residues. So, it is difficult to get only the most significant miRNA. So, a threshold value according to energy was taken as reference. miRNA which has the least energy value it will be the most significant miRNA because, miRNA with least energy value will have more interaction with ligands. The miRNA hsa-miR-1297 has the least energy value which is 27. So, this could be the significant miRNA and would be better to interact with ligands.
V Significant miRNA(has-miR-1297)
As discussed in the previous chapter the miRNA hsa-miR-1297 lies in the fourth family hsa-miR-26b-5p/miR-26-5p/1297/4465, this family has four members in which has-miR-1297 is the significant miRNA. Hsa-miR-1297 has up regulation in the liver cancer. While glioma, lung, adenocarcinoma and prostate cancer has down regulation. The most important factor is that it has the least binding energy among all the miRNAs of each family.
VI Structural Details
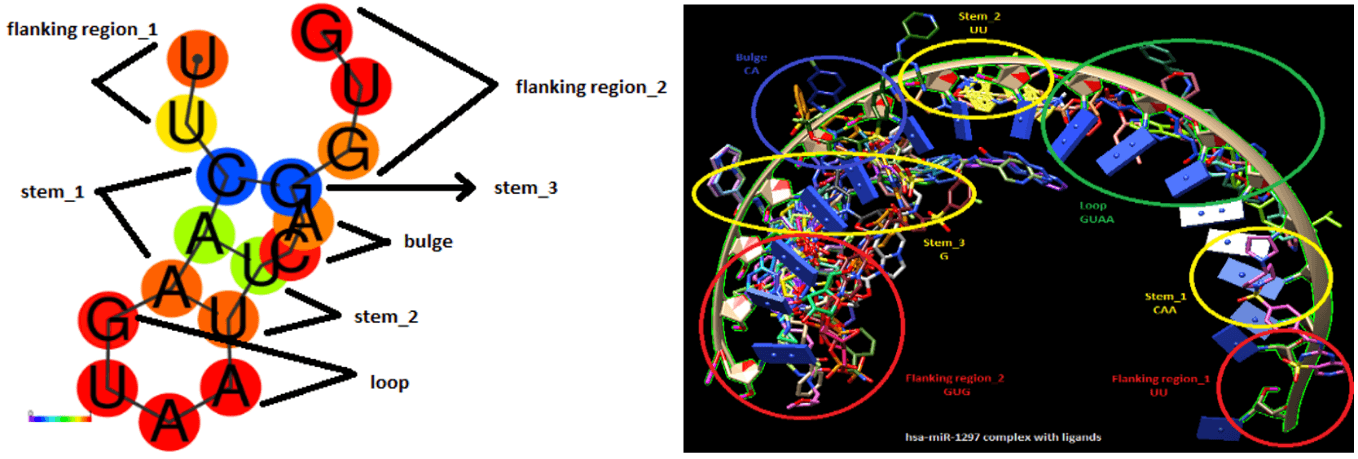
The miR-hsa-1297 has a sequence of 17 nucleotide bases. The structure comprises of 2 flanking regions, 3 stems, 1 loop and 1 bulge. The flanking region lies in the start from region 1-2 and at the end from 15-17. The stem regions are at 3-5,10-11 and at 14 positions while bulge at 12-13 positions. Loop forms at a region from 6-9. The (Figure 4A) shows the secondary structure of hsa-miR-1297. The (Figure 4B) shows the 3D structure viewed by chimera. It shows the positions of reported possible ligands in literature that bind to miRNA, has-miR-1297.
Figure 4: A) Secondary structure of hsa-miR-1297. B) 3D visualization of has-miR-1297 and its interaction with ligands.
Conclusion
This study focuses on microRNA's taking part in regulation of CDK6 gene and involved in Cancer. miRNA, hsa-miR-1297 belongs to the family 4 (hsa-miR-26b-5p/miR-26-5p/1297/4465), this family has four members (miRNAs), miRNA, hsa-miR-1297 has the least binding energy within the family and across the families. Hsa-miR-1297 is the most stable according to energy values to bind with ligands. The ligands are obtained from the PDB. The purpose of studying miRNAs in silico is to know the binding sites, behaviour, structure and the most important is the interaction with their targets. From the above study we came to know that miRNAs can be used as a drug targets, because it has high binding affinity towards their ligands.
Conflicts of Interest
None.
Article Info
Article Type
Research ArticlePublication history
Received: Fri 28, Aug 2020Accepted: Mon 07, Sep 2020
Published: Sat 19, Sep 2020
Copyright
© 2023 Sidra Batool. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.JSO.2020.04.11
Author Info
Muhammad Sibte Hasan Mahmood Tiyyaba Furqan Sidra Batool
Corresponding Author
Sidra BatoolDepartment of Biosciences, COMSATS University, Chak Shahzad, Islamabad, Pakistan
Figures & Tables
Table 1: Important features of CDK6 gene.
|
Feature keys |
Positions |
Descriptions |
|
Protein Kinase Domain |
13-300 |
CDK6 |
|
Binding site |
43 |
ATP |
|
Active site |
145 |
Proton Acceptor |
|
Nucleotide binding |
19-27 |
ATP |
|
Chain |
1-326 |
CDK6 |
Table 2: miRNAs with their families and involved in different pathways.
|
Sr # |
Family |
Members(miRNAs) |
Pathways |
|
1 |
hsa-miR-1305/hsa-miR-1305 |
hsa-miR-1305 |
P53 signaling pathway Viral carcinogenesis Non-small cell lung Pathways in cancer Chronic myeloid Hepatitis B Melanoma Pancreatic Leukemia Cell cycle Measles Glioma Cancer |
|
2 |
hsa-miR-548c-5p/ miR-548-5p/559
|
hsa-miR-548a-5p/hsa-miR-548ab/vhsa-miR-548ad-5p/hsa-miR-548ae-5p/hsa-miR-548ak/hsa-miR-548am-5p/hsa-miR-548ap-5p/ hsa-miR-548aq-5p/ hsa-miR-548ar-5p/hsa-miR-548as-5p/hsa-miR-548au-5p/hsa-miR-548ay-5p/hsa-miR-548b-5p/hsa-miR-548bb-5p/ hsa-miR-548c-5p/hsa-miR-548d-5p/hsa-miR-548h-5p/hsa-miR-548i/hsa-miR-548j-5p/hsa-miR-548o-5p/hsa-miR-548w/hsa-miR-548y/hsa-miR-559 |
|
|
3 |
hsa-miR-4429 /miR-320/4429 |
hsa-miR-320a/hsa-miR-320b/has-miR-320c/hsa-miR-320d/hsa-miR-4429 |
|
|
4 |
hsa-miR-26b-5p/miR-26-5p/1297/4465 |
hsa_miR-26a-5p/hsa_miR-26b-5p/hsa_miR-1297/hsa_miR-4465 |
|
|
5 |
hsa-miR-548k/miR-548-5p/8054 |
hsa_miR-548av-5p/hsa_miR-548k/hsa_miR-8054 |
|
|
6 |
hsa-miR-107/miR103-3P/107 |
hsa-miR-103a-3p/hsa-miR-107 |
|
|
7 |
hsa-miR-21-3p/miR-21-3p/3591-p |
hsa-miR-21-3p/hsa-miR3-591-3p |
|
|
8 |
hsa-miR-590-3p/miR-590-3p |
has-miR-590-3p |
|
|
9 |
hsa-miR-362-3p/miR-329-3p/362-3p |
hsa-miR-329-3p/has-miR-362-3p |
|
|
10 |
hsa-miR-335-3p/miR-335-5p |
hsa-miR-335-3p |
|
|
11 |
hsa-miR-33a-5p/miR-33-5p |
hsa-miR33a-5p/hsa-miR33b-5p |
|
|
12 |
hsa-miR-3140-3p/miR-3140-3p |
hsa-miR-3140-3p |
|
|
13 |
hsa-miR-3619-5p/miR-214-3p/761/3619-5p |
hsa-miR-214-3p/has-miR-761/hsa-miR-3619-5p |
|
|
14 |
hsa-miR-32-3p/miR-32-3p |
hsa-miR-32-3p |
|
|
15 |
hsa-miR-548e-3p/miR-548ar-3p/548ar-3p/548az-3p/548e-3p/548f-3p |
hsa-miR-548a-3p/hsa-miR-548ar-3p/hsa-miR-548az-3p/hsa-miR-548e-3p/hsa-miR-548f-3p |
|
|
16 |
hsa-miR-576-5p/miR-576-5p |
hsa-miR-576-5p |
|
|
17 |
hsa-miR-548o-3p/miR-548-3p/1323 |
hsa-miR-548o-3p/hsa-miR-1323 |
|
|
18 |
hsa-miR-320e/miR-320 |
hsa-miR-320e |
|
|
19 |
hsa-miR-424-5p/miR-15-5p/16-5p/195-5p/424-5p/497-5p/6838-5p |
hsa-miR-15a-5p/hsa-miR-15b-5p/hsa-miR-16-5p/hsa-miR-195-5p/has-miR-424-5p/hsa-miR-497-5p/hsa-miR-6838-5p |
|
|
20 |
hsa-miR-450b-5p/miR-450-5p |
hsa-miR-450b-5p |
|
|
21 |
hsa-miR-142-5p/miR-142-5p/5590-3p |
hsa-miR-142-5p/hsa-miR-5590-3p |
|
|
22 |
hsa-miR-29b-3p/miR-29-3p |
hsa-miR-29a-3p/hsamiR-29b-3p/hsa-miR-29c-3p |
|
|
23 |
hsa-miR-589-3p/miR-589-3p |
hsa-miR-589-3p |
Table 3: Different regions present in secondary structure.
|
Sr # |
Family |
Members(miRNAs) |
Flanking Regions |
Stems |
Loops |
Bulges |
Seed Region |
|
1 |
hsa-miR-1305/ hsa-miR-1305 |
hsa-miR-1305 |
1-7/20-22 |
8-10/17-19 |
11-16 |
- |
UUUCAAC
|
|
2 |
hsa-miR-548c-5p/ miR-548-5p/559
|
a) hsa-miR-548a-5p |
1-2/11-22 |
3-4/9-10 |
5-8 |
- |
AAAGUAA |
|
b) hsa-miR-548ab |
1-2/22 |
3-4/9-13/9-21 |
5-8/14-18 |
- |
|||
|
c) hsa-miR-548ad-5p |
1-2/11-20 |
3-4/9-10 |
5-8 |
- |
|||
|
d) hsa-miR-548ae-5p |
1-2/11-20 |
3-4/9-10 |
5-8 |
- |
|||
|
e) hsa-miR-548ak |
1-6/17-21 |
7-9/14-16 |
10-13 |
- |
|||
|
f) hsa-miR-548am-5p |
1-2 |
3-4/9-10/13-15/20-22 |
5-8/16-19 |
11-12 |
|||
|
g) hsa-miR-548ap-5p |
1-2/11-19 |
3-4/9-10 |
5-8 |
- |
|||
|
h) hsa-miR-548aq-5p |
1-2/22 |
3-4/9-12/20-21 |
5-8/13-19 |
- |
|||
|
i) hsa-miR-548ar-5p |
1-2 |
3-4/9-13/19-21 |
5-8/14-18 |
- |
|||
|
j) hsa-miR-548as-5p |
1-2/22 |
3-4/9-12/20-21 |
5-8/13-19 |
- |
|||
|
k) hsa-miR-548au-5p |
1-2 |
3-4/9-12/20-21 |
5-8/13-19 |
- |
|||
|
l) hsa-miR-548ay-5p |
1-2/11-21 |
3-4/9-10 |
5-8 |
- |
|||
|
m) hsa-miR-548b-5p |
1-2 |
3-4/9-10/13-15/20-22 |
5-8/16-19 |
11-12 |
|||
|
n) hsa-miR-548bb-5p |
1-12 |
13-15/20-22 |
16-19 |
- |
|||
|
o) hsa-miR-548c-5p |
1-2 |
3-4/9-10/13-15/20-24 |
5-8/16-19 |
11-12 |
|||
|
p) hsa-miR-548d-5p |
1-2 |
3-4/9-10/13-15/20-22 |
5-8/16-19 |
11-12 |
|||
|
q) hsa-miR-548h-5p |
1-3/10-22 |
4/9 |
5-8 |
- |
|||
|
r) hsa-miR-548i |
1-2/22 |
3-4/9-12/20-21 |
5-8/13-19 |
- |
|||
|
s) hsa-miR-548j-5p |
1-2/11-22 |
3-4/9-10 |
5-8 |
- |
|||
|
t) hsa-miR-548o-5p |
1-2 |
3-4/9-10/13-15/20-22 |
5-8/16-19 |
11-12 |
|||
|
u) hsa-miR-548w |
1-4/23 |
5/12-15/20-22 |
6-11/16-19 |
- |
|||
|
v) hsa-miR-548y |
1-3/22 |
4/9/14-15/20-21 |
5-8/16-19 |
10-13 |
|||
|
w) hsa-miR-559 |
1-4/15-21 |
5-7/12-14 |
8-11 |
- |
|||
|
3 |
hsa-miR-4429/miR-320/4429 |
hsa-miR-320a |
1-8/21-22 |
9/11-12/17-18/20 |
13-16 |
10/19 |
AAAGCUG
|
|
a) hsa-miR-320b |
1-8/21-22 |
9/11-12/17-18/20 |
13-16 |
10/19 |
|||
|
b) has-miR-320c |
1-10/19-20 |
11-12/17-18 |
13-16 |
- |
|||
|
c) hsa-miR-320d |
1-10/19 |
11-12/17-18 |
13-16 |
- |
|||
|
d) hsa-miR-4429 |
1-9/20 |
10-12/17-19 |
13-16 |
- |
|||
|
4 |
hsa-miR-26b-5p/miR-26-5p/1297/4465 |
a) hsa_miR-26a-5p |
1-4/21 |
5/10-12/19-20 |
6-9/13-18 |
- |
UCAAGUA
|
|
b) hsa_miR-26b-5p |
1-2/15-21 |
3-5/10-11/14 |
6-9 |
12-13 |
|||
|
c) hsa_miR-1297 |
1-2/15-17 |
3-5/10-11/14 |
6-9 |
12-13 |
|||
|
d) hsa_miR-4465 |
1-2/14-22 |
3/6/11/13 |
7-10 |
4-5/12 |
|||
|
5 |
hsa-miR-548k/miR-548-5p/8054 |
a) hsa_miR-548av-5p |
1-7/14-18 |
8/10-11/13 |
--- |
- |
AAAGUAC |
|
b) hsa_miR-548k |
1-10/22 |
11-12/20-21 |
13-19 |
- |
|||
|
c)hsa_miR-8054 |
1-7/19-21 |
8-9/17-18 |
10-16 |
- |
|||
|
6 |
hsa-miR-107/miR103-3P/107 |
a) hsa-miR-103a-3p |
1-3/20-23 |
4-6/17-19 |
7-16 |
- |
GCAGCAU |
|
b) hsa-miR-107 |
1-3/20-23 |
4-6/17-19 |
7-16 |
- |
|||
|
7 |
hsa-miR-21-3p/miR-21-3p/3591-p |
a) hsa-miR-21-3p |
1-6/21 |
7-11/16-20 |
12-15 |
- |
AACACCA |
|
b) has-miR-3591-3p |
1-312-22 |
4-5/10-11 |
6-9 |
- |
|||
|
8 |
hsa-miR-590-3p/miR-590-3p |
has-miR-590-3p |
1-9/18-21 |
10/17 |
11-16 |
- |
AAUUUUA |
|
9 |
hsa-miR-362-3p/miR-329-3p/362-3p |
a) hsa-miR-329-3p |
1-3/18-22 |
4/9-11/16-17 |
5-8/12-15 |
- |
ACACACC |
|
b) has-miR-362-3p |
1-6/18-22 |
7-9/15-17 |
10-14 |
- |
|||
|
10 |
hsa-miR-335-3p/miR-335-5p |
hsa-miR-335-3p |
1-4/21-22 |
5-7/18-20 |
8-17 |
- |
UUUUCAU |
|
11 |
hsa-miR-33a-5p/miR-33-5p
|
a) hsa-miR33a-5p |
1-2/21 |
3-5/7-9/14-16/18-20 |
10-13 |
6/17 |
UGCAUUG |
|
b) hsa-miR33b-5p |
1-2 |
3-5/7-9/14-16/18-20 |
10-13 |
6/17 |
|||
|
12 |
hsa-miR-3140-3p/miR-3140-3p |
hsa-miR-3140-3p |
1/20-22 |
2-5/16-19 |
6-15 |
- |
GCUUUUG |
|
13 |
hsa-miR-3619-5p/miR-214-3p/761/3619-5p |
a) hsa-miR-214-3p |
1-3/20-22 |
4-5/18-19 |
6-17 |
- |
CAGCAGG
|
|
b) has-miR-761 |
1-4/18-22 |
5-7/15-17 |
8-14 |
- |
|||
|
c) hsa-miR-3619-5p |
1/16-22 |
2-5/12-15 |
6-11 |
- |
|||
|
14 |
hsa-miR-32-3p/miR-32-3p |
hsa-miR-32-3p |
- |
- |
1-22 |
- |
AAUUUAG |
|
15 |
hsa-miR-548e-3p/miR-548ar-3p/548ar-3p/548az-3p/548e-3p/548f-3p |
a) hsa-miR-548a-3p |
1-8 |
9-12/19-22 |
13-18 |
- |
AAAACUG |
|
b) hsa-miR-548ar-3p |
1-7 |
8-11/18-21 |
12-17 |
- |
|||
|
c) hsa-miR-548az-3p |
1-7 |
8-11/18-21 |
12-17 |
- |
|||
|
d) hsa-miR-548e-3p |
- |
- |
1-22 |
- |
|||
|
e) hsa-miR-548f-3p |
- |
- |
1-19 |
- |
|||
|
16 |
hsa-miR-576-5p/miR-576-5p |
hsa-miR-576-5p |
- |
- |
1-22 |
- |
UUCUAAU |
|
17 |
hsa-miR-548o-3p/miR-548-3p/1323 |
a) hsa-miR-548o-3p |
1/22 |
2-6/17-21 |
7-16 |
|
CAAAACU |
|
b) hsa-miR-1323 |
- |
- |
1-22 |
- |
|||
|
18 |
hsa-miR-320e/miR-320 |
hsa-miR-320e |
1-4/18 |
5-6/16-17 |
7-15 |
- |
AAGCUGG |
|
19 |
hsa-miR-424-5p/miR-15-5p/16-5p/195-5p/424-5p/497-5p/6838-5p |
a) hsa-miR-15a-5p |
1-6 |
7-10/19-22 |
11-18 |
- |
AGCAGCA |
|
b) hsa-miR-15b-5p |
1-3/16-22 |
4-5/14-15 |
6-13 |
- |
|||
|
c) hsa-miR-16-5p |
1-5/22 |
6-7/20-21 |
8-19 |
- |
|||
|
d) hsa-miR-195-5p |
1-2 |
3-4/10/15/20-21 |
11-14 |
5-9/16-19 |
|||
|
e) has-miR-424-5p |
1-4/21-22 |
5/10/12-13/19-20 |
69/14-18 |
11 |
|||
|
f) hsa-miR-497-5p |
1-2/15-21 |
3-6/11-14 |
7-10 |
- |
|||
|
g) hsa-miR-6838-5p |
1-2/15-22 |
3-4/12-13 |
5-11 |
|
|||
|
20 |
hsa-miR-450b-5p/miR-450-5p |
hsa-miR-450b-5p |
- |
- |
- |
1-22 |
UUUGCAA
|
|
21 |
hsa-miR-142-5p/ miR-142-5p/5590-3p |
a) hsa-miR-142-5p |
1-5 |
6-10/17-21 |
11-16 |
- |
AUAAAGU
|
|
b) hsa-miR-5590-3p |
1-6/20-21 |
7-9/17-19 |
10-16 |
- |
|||
|
22 |
hsa-miR-29b-3p/miR-29-3p |
a) hsa-miR-29a-3p |
1-4/21-22 |
5-7/18-20 |
8-17 |
- |
AGCACCA
|
|
b) hsamiR-29b-3p |
1-2/22-23 |
3-6/19-21 |
7-18 |
- |
|||
|
c) hsa-miR-29c-3p |
1-4/21-22 |
5-7/18-20 |
18-17 |
- |
|||
|
23 |
hsa-miR-589-3p/miR-589-3p |
hsa-miR-589-3p |
1-3/20-24 |
4-7/16-19 |
8-15 |
- |
CAGAACA |
Table 4: Mapping of CDK6 residues and miRNAs.
|
Sr # |
Family |
miRNA |
Energy |
Protein Residues Mapping + mRNA Mappiing |
miRNA Residues Mapping |
|
1 |
hsa-miR-1305/ hsa-miR-1305
|
hsa-miR-1305 |
-0.16 |
ARG90 = AGA + PK-Domain ARG87 = CGA + PK-Domain ARG78 = AGG + PK-Domain ARG186 = UAC + PK-Domain GLU72 = GAG + PK-Domain PHE135 = GAC + PK-Domain TYR292 = UCU + PK-Domain VAL181 = GUC + PK-Domain ARG144 = CAU + PK-Domain ALA167 = CUU + PK-Domain LEU166 = GGC + PK-Domain GLY165 = UUC + PK-Domain GLU21 = GAG + PK-Domain SER195 = UCC + PK-Domain SER194 = CAG + PK-Domain GLN193 = CUC + PK-Domain |
UUUUCAACU A=6,7 U=1,2,3,4 C=5,8 |
|
2 |
hsa-miR-548c-5p/ miR-548-5p/559 |
hsa-miR-548ap-5P |
-7.46 |
ARG46 = CGG + PK-Domain TYR24 = UAU + PK-Domain + NB-ATP LEU96 = UUA + PK-Domain GLN193 = CAG + PK-Domain SER195 = CUA + PK-Domain TYR196 = CGC + PK-Domain ARG144 = CGC + PK-Domain PRO244 = UAG + PK-Domain |
AAAAGUAAU A=2,3,4,7,8 G=5 U=6 |
|
3 |
hsa-miR-4429/miR-320/4429 |
hsa-miR-320b |
-1.2 |
VAL179 = GUG + PK-Domain ILE169 = AUC + PK-Domain SER171 = AGU + PK-Domain TYR170 = UAU + PK-Domain TRP184 = UGG + PK-Domain GLN103 = CAA + PK-Domain TYR185 = UAC + PK-Domain ARG168 = CGC + PK-Domain GLN149 = CAG + PK-Domain PRO17 = GCG + PK-Domain LYS216 = GCC + PK-Domain LEU152 = CUG + PK-Domain LEU166 = CUU + PK-Domain GLU18 = GAG + PK-Domain GLY20 = GGG + PK-Domain + NB-ATP ILE19 = AUC + PK-Domain + NB-ATP ALA17 = GCG + PK-Domain LYS111 = AAA + PK-Domain PHE164 = UUC + PK-Domain ASN150 = AAC + PK-Domain PRO148 = CCA + PK-Domain THR106 = ACC + PK-Domain ILE151 = AUU + PK-Domain ASP110 = GAU + PK-Domain HIS100 = CAU + PK-Domain ASP104 = GAC + PK-Domain LEU105 = UUG + PK-Domain VAL101 = GUC + PK-Domain ASP102 = GAU + PK-Domain |
AAAAGCUGGG A=2,3,4 G=5,8 C=6 U=7 |
|
4 |
hsa-miR-26b-5p/miR-26-5p/1297/4465
|
hsa-miR-1297 |
-10.63 |
GLU21 = GAG + PK-Domain + NB-ATP LEU166 = CUU + PK-Domain ALA167 = GCC + PK-Domain THR107 = ACU + PK-Domain ASP110 = GAU + PK-Domain LYS111 = AAA + PK-Domain TYR196 = CGC + PK-Domain ALA197 = CAC + PK-Domain ARG140 = CGA + PK-Domain THR198 = CCC + PK-Domain HIS137 = CAU + PK-Domain SER138 = UCA + PK-Domain |
UUCAAGUAA A=4,5,8 G=6 U=2,7 C=3 |
|
5 |
hsa-miR-548k/miR-548-5p/8054 |
hsa-miR-548av-5p |
-6.29 |
ALA167 = GCC + PK-Domain GLN193 = CAG + PK-Domain ILE169 = AUC + PK-Domain VAL181 = GUC + PK-Domain ARG186 = AGA + PK-Domain ARG78 = AGG + PK-Domain HIS139 = CAC + PK-Domain GLU72 = GAG + PK-Domain SER139 = CAC + PK-Domain PHE135 = UUU + PK-Domain TYR292 = CAG + PK-Domain |
AAAAGUACU A=2,3,4,7 G=5 U=6 C=8 |
|
6 |
hsa-miR-107/miR103-3P/107 |
hsa-miR-107 |
-1.2 |
VAL179 = GUG + PK-Domain GLN193 = CAG + PK-Domain SER155 = AGC + PK-Domain SER156 = AGC + PK-Domain TYR108 = UAC + PK-Domain LYS111 = AAA + PK-Domain VAL112 = GUU + PK-Domain THR121 = ACC + PK-Domain PRO113 = CCA + PK-Domain ARG140 = CGA + PK-Domain LEU94 = CUA + PK-Domain THR84 = ACA + PK-Domain THR92 = ACC + PK-Domain VAL47 = GUG + PK-Domain PRO118 = CCC + PK-Domain |
AGCAGCAUU A=4,7 G=2,5 U=8 C=3,6
|
|
7 |
hsa-miR-21-3p/miR-21-3p/3591-p
|
hsa-miR-3591-3p |
-5.8 |
GLN193 = CAG + PK-Domain SER194 = UAG + PK-Domain SER195 = CUA + PK-Domain ARG144 = CGC + PK-Domain ALA197 = CAC + PK-Domain TYR292 = CAG + PK-Domain |
AAACACCAU A=2,3,5,8 G= -- U= -- C=4,6,7 |
|
8 |
hsa-miR-590-3p/miR-590-3p
|
hsa-miR-590-3p |
-0.93 |
ARG140 = CGA + PK-Domain SER195 = CUA + PK-Domain ALA197 = CAC + PK-Domain ASN284 = CCC + PK-Domain ALA280 = UUU + PK-Domain ARG78 = AGG + PK-Domain GLY157 = GGA + PK-Domain SER156 = AGC + PK-Domain THR121 = ACC + PK-Domain PRO118 = CCC + PK-Domain GLU120 = GAA + PK-Domain |
UAAUUUUAU A=2,3,8 G= U=4,5,6,7 C=
|
|
9 |
hsa-miR-362-3p/miR-329-3p/362-3p
|
hsa-miR-329-3P |
-2.64 |
GLU21 = GAG + PK-Domain +NB-ATP LEU166 = CUU + PK-Domain ALA167 = GCC + PK-Domain LYS111 = AAA + PK-Domain PRO113 = CCA + PK-Domain GLN193 = CAG + PK-Domain ARG144 = CGC + PK-Domain SER195 = CUA + PK-Domain VAL142 = GUG + PK-Domain TYR196 = CGC + PK-Domain ALA197 = CAC + PK-Domain THR198 = CCC + PK-Domain HIS137 = CAU + PK-Domain SER138 = UCA + PK-Domain ARG140 = CGA + PK-Domain |
AACACACCU A=2,4,6, G= U= C=3,5,7,8 |
|
10 |
hsa-miR-335-3p/miR-335-5p
|
hsa-miR-335-3p |
0.12 |
SER194 = UAG + PK-Domain ASP246 = UGU + PK-Domain GLN193 = CAG + PK-Domain VAL179 = GUG + PK-Domain ILE169 = AUC + PK-Domain TYR292 = CAG + PK-Domain |
UUUUUCAUU A=7 G= U=2,3,4,5,8 C=6 |
|
11 |
hsa-miR-33a-5p/miR-33-5p
|
hsa-miR33a-5p |
-0.81 |
ASP246 = UGU + PK-Domain GLN193 = CAG + PK-Domain SER194 = UAG + PK-Domain SER195 = CUA + PK-Domain VAL190 = GUC + PK-Domain ARG144 = CGC + PK-Domain VAL181 = GUC + PK-Domain GLU189 = GAA + PK-Domain ALA197 = CAC + PK-Domain ALA23 = GCC + PK-Domain + NB-ATP GLY22 = GGC + PK-Domain + NB-ATP ARG140 = CGA + PK-Domain VAL142 = GUG + PK-Domain VAL141 = GUA + PK-Domain HIS143 = CAU + PK-Domain |
GUGCAUUGUAGUUGCAUUGCA A=5,16 G=3,8,14,19 U=2,6,7,13,17,18 C=4,15
|
|
12 |
hsa-miR-3140-3p/miR-3140-3p
|
hsa-miR-3140-3p |
-6.79 |
VAL16 = GUG + PK-Domain VAL17 = GCG + PK-Domain PRO74 = CCC + PK-Domain TYR292 = CAG + PK-Domain SER155 = AGC + PK-Domain SER156 = AGC + PK-Domain SER293 = UGC + PK-Domain LYS279 = GUG + PK-Domain SER290 = UGC + PK-Domain LYS111 = AAA + PK-Domain ALA286 = CAA + PK-Domain LYS287 = AAG + PK-Domain |
AGCUUUUGG A= G=2,8 U=4,5,6,7 C=3
|
|
13 |
hsa-miR-3619-5p/miR-214-3p/761/3619-5p
|
hsa-miR-214-3p |
-2.26 |
THR84 = ACA + PK-Domain LEU94 = CUA + PK-Domain VAL82 = GUG + PK-Domain ALA23 = GCC + PK-Domain + NB-ATP TYR24 = UAU + PK-Domain + NB-ATP GLY22 = GGC + PK-Domain + NB-ATP ARG140 = CGA + PK-Domain VAL142 = GUG + PK-Domain VAL141 = GUA + PK-Domain ARG144 = CGC + PK-Domain TYR196 = CGC + PK-Domain ARG186 = AGA + PK-Domain VAL181 = GUC + PK-Domain ILE169 = AUC + PK-Domain LEU166 = CUU + PK-Domain ALA167 = GCC + PK-Domain LYS111 = AAA + PK-Domain TYR108 = UAC + PK-Domain VAL112 = GUU + PK-Domain GLY116 = GGA + PK-Domain PRO113 = CCA + PK-Domain PRO115 = CCU + PK-Domain PRO118 = CCC + PK-Domain VAL117 = GUG + PK-Domain THR119 = ACU + PK-Domain |
ACAGCAGGC A=3,6 G=4,7,8 U= C=2,5 |
|
14 |
hsa-miR-32-3p/miR-32-3p
|
hsa-miR-32-3p |
-5.18 |
SER194 = UAG + PK-Domain ASP246 = CUA + PK-Domain GLN193 = CAG + PK-Domain LEU166 = CUU + PK-Domain ALA167 = GCC + PK-Domain ILE169 = AUC + PK-Domain ARG168 = CGC + PK-Domain TYR292 = CAG + PK-Domain |
CAAUUUAGU A=2,3,7 G=8 U=4,5,6 C= |
|
15 |
hsa-miR-548e-3p/miR-548ar-3p/548ar-3p/548az-3p/548e-3p/548f-3p
|
hsa-miR-548e-3p |
-6.04 |
GLN193 = CAG + PK-Domain SER194 = UAG + PK-Domain SER195 = CUA + PK-Domain TYR196 = CGC + PK-Domain ALA197 = CAC + PK-Domain THR198 = CCC + PK-Domain ASP110 = GAU + PK-Domain LYS111 = AAA + PK-Domain |
AAAAACUGA A=2,3,4,5 G=8 U=7 C=6 |
|
16 |
hsa-miR-576-5p/ miR-576-5p
|
hsa-miR-576-5p |
-0.47 |
SER19 = AUC + PK-Domain TYR196 = CGC + PK-Domain ALA197 = CAC + PK-Domain ILE169 = AUC + PK-Domain |
AUUCUAAUU A=6,7 G= U=2,3,5,8 C=4 |
|
17 |
hsa-miR-548o-3p/miR-548-3p/1323 |
hsa-miR-1323 |
0.16 |
ARG140 = CGA + PK-Domain TYR196 = CGC + PK-Domain VAL142 = GUG + PK-Domain HIS137 = CAU + PK-Domain ALA23 = GCC + PK-Domain + NB-ATP GLU21 = GAG + PK-Domain + NB-ATP LEU166 = CUU + PK-Domain GLY165 = GGC + PK-Domain ARG168 = CGC + PK-Domain GLN149 = CAG + PK-Domain ASP110 = GAU + PK-Domain LYS216 = GCC + PK-Domain ARG215 = AAA + PK-Domain GLU114 = GAG + PK-Domain THR267 = AGA + PK-Domain |
CCAAAACUG A=3,4,5,6 G= U=8 C=2,7 |
|
18 |
hsa-miR-320e/miR-320
|
hsa-miR-320e |
-9.15 |
LEU166 = CUU + PK-Domain ALA167 = GCC + PK-Domain ILE169 = AUC + PK-Domain LYS111 = AAA + PK-Domain PRO113 = CCA + PK-Domain PRO118 = CCC + PK-Domain ALA23 = GCC + PK-Domain + NB-ATP ARG144 = CGC + PK-Domain SER195 = CUA + PK-Domain VAL142 = GUG + PK-Domain TYR196 = CGC + PK-Domain THR198 = CCC + PK-Domain ARG140 = CGA + PK-Domain SER138 = UCA + PK-Domain |
AAAGCUGGG A=2,3 G=4,7,8 U=6 C=5 |
|
19 |
hsa-miR-424-5p/miR-15-5p/16-5p/195-5p/424-5p/497-5p/6838-5p |
hsa-miR-15b-5p |
-5.65 |
VAL16 = GUG + PK-Domain TYR24 = UAU + PK-Domain + NB-TP ARG44 = CGC + PK-Domain ARG186 = AGA + PK-Domain ASP145 = GAU + PK-Domain HIS143 = CAU + PK-Domain ARG78 = AGG + PK-Domain GLU72 = GAG + PK-Domain LYS111 = AAA + PK-Domain SER155 = AGC + PK-Domain |
UAGCAGCAC A=2,5,8 G=3,6 U= C=4,7 |
|
20 |
hsa-miR-450b-5p/miR-450-5p |
hsa-miR-450b-5p |
-1.22 |
ARG46 = CGG + PK-Domain VAL47 = GUG + PK-Domain GLN48 = CAG + PK-Domain ALA23 = GCC + PK-Domain LEU94 = CUA + PK-Domain TYR24 = UAU + PK-Domain + NB-ATP VAL82 = GUG + PK-Domain LEU96 = UUA + PK-Domain ARG144 = CGC + PK-Domain ARG186 = AGA + PK-Domain ASP145 = GAU + PK-Domain-Active Site LEU79 = UUG + PK-Domain ARG78 = AGG + PK-Domain GLU72 = GAG + PK-Domain VAL16 = GUG + PK-Domain LYS111 = AAA + PK-Domain THR154 = ACC + PK-Domain GLN18 = GAG + PK-Domain SER155 = AGC + PK-Domain GLY157 = GGA + PK-Domain SER156 = AGC + PK-Domain |
UUUUGCAAU A=7,8 G=5 U=2,3,4 C=6
|
|
21 |
hsa-miR-142-5p/ miR-142-5p/5590-3p |
hsa-miR-142-5p |
-3.36 |
VAL147 = AAA + PK-Domain GLN48 = CAG + PK-Domain VAL45 = GUG + PK-Domain LEU94 = CUA + PK-Domain TYR24 = UAU + PK-Domain +NB-ATP ASP246 = UGU + PK-Domain GLN193 = CAG + PK-Domain ALA248 = CCU + PK-Domain VAL82 = GUG + PK-Domain LEU96 = UUA + PK-Domain LYS43 = AAG + PK-Domain + ATP-Binding Site ASP163 = GAC + PK-Domain VAL247 = UGC + PK-Domain SER194 = CCU + PK-Domain LEU79 = UUG + PK-Domain VAL142 = GUG + PK-Domain ALA162 = GCU + PK-Domain ARG140 = CGA + PK-Domain HIS143 = CAU + PK-Domain LEU161 = CUC + PK-Domain VAL141 = GUA + PK-Domain LEU136 = CUU + PK-Domain VAL76 = GUG + PK-Domain |
CAUAAAGUA A=2,4,5,6 G=7 U=3,8 C=
|
|
22 |
hsa-miR-29b-3p/miR-29-3p |
hsa-miR-29c-3p |
-6.13 |
VAL47 = GUG + PK-Domain LEU94 = CUA + PK-Domain ALA23 = GCC + PK-Domain + NB-ATP GLU21 = GAG + PK-Domain + NB-ATP GLY22 = GGC + PK-Domain + NB-ATP GLY165 = GGC + PK-Domain VAL181 = GUC + PK-Domain SER223 = AGA + PK-Domain ASP224 = UGU + PK-Domain ARG144 = CGC + PK-Domain ARG186 = AGA + PK-Domain ASP145 = GAU + PK-Domain LYS147 = AAA + PK-Domain VAL82 = GUG + PK-Domain ARG140 = CGA + PK-Domain ASP242 = CUG + PK-Domain THR198 = CCC + PK-Domain PRO199 = CGU + PK-Domain PHE283 = UAA + PK-Domain PRO238 = AGG + PK-Domain ARG288 = AAU + PK-Domain THR282 = AUU + PK-Domain ASN284 = CCC + PK-Domain PRO285 = AGC + PK-Domain ALA286 = CAA + PK-Domain LYS287 = AAG + PK-Domain ILE289 = AUC + PK-Domain LEU237 = CCC + PK-Domain GLY236 = ACU + PK-Domain LYS279 = GUG + PK-Domain |
UAGCACCAU A=2,5,8 G=3 U= C=4,6,7 |
|
23 |
hsa-miR-589-3p/miR-589-3p |
hsa-miR-589-3p |
8.2 |
LEU94 = CUA + PK-Domain CYS83 = UGC + PK-Domain VAL82 = GUG + PK-Domain THR95 = ACU + PK-Domain ASP81 = GAU + PK-Domain LEU96 = UUA + PK-Domain VAL97 = GUG + PK-Domain LEU79 = UUG + PK-Domain GLN103 = CAA + PK-Domain VAL77 = GUC + PK-Domain ALA41 = GCG + PK-Domain PHE98 = UUU + PK-Domain PHE28 = UUC + PK-Domain LYS43 = AAG + PK-Domain + ATP-Binding Site LEU42 = UUG + PK-Domain ALA23 = GCC + PK-Domain + NB-ATP LYS26 = AAG + PK-Domain + NB-ATP THR106 = ACC + PK-Domain + NB-ATP GLU21 = GAG + PK-Domain + NB-ATP GLU18 = GAG + PK-Domain VAL27 = GUG + PK-Domain + NB-ATP ALA17 = GCG + PK-Domain GLY20 = GGG + PK-Domain + NB-ATP VAL142 = GUG + PK-Domain ILE19 = AUC + PK-Domain + NB-ATP PHE164 = UUC + PK-Domain THR107 = ACU + PK-Domain HIS143 = CAU + PK-Domain LYS29 = AAG + PK-Domain VAL77 = GUC + PK-Domain HIS100 = CAU + PK-Domain GLU99 = GAA + PK-Domain ASP102 = GAU + PK-Domain ASP145 = GAU + PK-Domain ASN150 = AAC + PK-Domain LYS111 = AAA + PK-Domain ARG78 = AGG + PK-Domain ARG144 = CGC + PK-Domain ARG186 = AGA + PK-Domain ALA167 = GCC + PK-Domain VAL181 = GUC + PK-Domain VAL190 = GUC + PK-Domain THR182 = GAU + PK-Domain LEU146 = CUA + PK-Domain ARG168 = CGC + PK-Domain LYS147 = AAA + PK-Domain ILE169 = AUC + PK-Domain TYR185 = UAC + PK-Domain GLN149 = CAG + PK-Domain PRO148 = CCA + PK-Domain SER204 = GAG + PK-Domain ASP104 = GAC + PK-Domain GLN193 = CAG + PK-Domain |
UCAGAACAA A=3,5,6,8 G=4 U= C=2,7
|
References
- Natascha Bushati, Stephen M Cohen (2007) microRNA functions. Annu Rev Cell Dev Biol 23: 175-205. [Crossref]
- Milena S Nicoloso, Riccardo Spizzo, Masayoshi Shimizu, Simona Rossi, George A Calin (2009) MicroRNAs-the micro steering wheel of tumour metastases. Nat Rev Cancer 9: 293-302. [Crossref]
- Aurora Esquela Kerscher, Frank J Slack (2006) Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6: 259-269. [Crossref]
- Jun Lu, Shangqin Guo, Benjamin L Ebert, Hao Zhang, Xiao Peng et al. (2008) MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell 14: 843-853. [Crossref]
- Carsten Detering, Gabriele Varani (2004) Validation of automated docking programs for docking and database screening against RNA drug targets. J Med Chemist 47: 4188-4201. [Crossref]
- Nicolas Moitessier, Eric Westhof, Stephen Hanessian (2006) Docking of aminoglycosides to hydrated and flexible RNA. J Med Chem 49: 1023-1033. [Crossref]
- Patrick Pfeffer, Holger Gohlke (2007) DrugScoreRNA--Knowledge-Based Scoring Function To Predict RNA−Ligand Interactions. J Chem Inf Model 47: 1868-1876. [Crossref]
- Marcin Biesiada, Katarzyna J Purzycka, Marta Szachniuk, Jacek Blazewicz, Ryszard W Adamiak (2016) Automated RNA 3D Structure Prediction with RNAComposer. Methods Mol Biol 1490: 199-215. [Crossref]
- Chih Hung Chou, Nai Wen Chang, Sirjana Shrestha, Sheng Da Hsu, Yu Ling Lin et al. (2016) miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 44: D239-D247. [Crossref]
- Garrett M Morris, Ruth Huey, William Lindstrom, Michel F Sanner, Richard K Belew et al. (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30: 2785-2791. [Crossref]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE et al. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Computational Chemistry 19:1639-1662.
- Roman A Laskowski, Mark B Swindells (2011) LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J Chem Inf Model 51: 2778-2786. [Crossref]
- Eric F Pettersen, Thomas D Goddard, Conrad C Huang, Gregory S Couch, Daniel M Greenblatt et al. (2004) UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 25: 1605-1612. [Crossref]
