Liquid Biopsy in Early Breast Cancer: A Preliminary Report
A B S T R A C T
Background: Liquid biopsy (LB) is a technique that utilizes circulating biomarkers from cancer patients to provide information regarding the genetic landscape of the cancer. LB is emerging as an alternative and complementary diagnostic and prognostic tool to surgical biopsy and is expected to provide the tool for the implementation of precision oncology in clinical settings. In fact, it may contribute to enhance understanding of tumor heterogeneity and permitting the dynamic monitoring of treatment responses and genomic variations. Thus, LB is a promising method for the management of cancer, including breast cancer (BC), whose incidence in Italy is progressively increasing. Previous studies focused mainly on patients with advanced-stage BC. In the present study we evaluated the number of circulating tumor cells (CTCs), the quantity of cell free tumor DNA (cftDNA) and the analysis of the mutational profile of DNA from CTCs (ctcDNA) and cftDNA in early stage BC patients.
Methods: Matched pre- and post-surgery blood samples were collected from 47 early stage BC patients. CTCs enumeration was done using Isoflux system, molecular profile of ctcDNA and cftDNA was performed with the Spotlight 59 Panels kit on a MiSeq Illumina instrument.
Results: Eighty percent of samples was CTCs-positive, while healthy controls were all CTCs-negative. Forty-four patients provided a pre-surgery and 21 post-surgery sample. By comparing the number of CTCs post-surgery with that of pre-surgery, we found that 66% of patients showed a decreased number of CTCs, 14% of patients continued to have the same number of CTCs, while, interestingly, 19% of patients showed an increased number of CTCs. Next Generation Sequencing (NGS) of ctcDNA and cftDNA showed that 52% of samples had mutations in 9 genes (TP53, CDKN2A, FBXW7, PTPN11, KRAS, NRAS, BRAF, IDH1, ALK) and in 5 genes (PIK3CA, APC ALK, KRAS, TSC1), respectively, with KRAS and ALK overlapping and TP53 being the most frequently mutated gene in ctcDNA analysis.
Conclusions: LB could facilitate early detection of minimal residual disease, aiding in the initiation of adjuvant therapy to prevent recurrence and progression towards metastasis, enhance individualized treatment and longitudinal screening, thus improving the clinical management and outcome of patients with early BC.
Keywords
Early breast cancer, liquid Biopsy, circulating tumor cells, cell-free tumor DNA, next generation sequencing
Background
Breast cancer (BC) is an invalidating disease with a serious physical and psychological impact. In Italy, increasing trends in BC incidence have been observed, so that today it represents the most common tumor (52,800 new cases in 2018), overcoming other major malignancies, such as colon and lung cancers (the so called “big killers”) [1]. Alarmingly, 41% of the new diagnosed tumors applies to more and more young people (age < 50 years), a percentage out of the age expected for diagnostic mammographic screening (age > 50 years). The first cause of death in cancer patients is the metastatic spread of the tumor from the primary site. As known, this occurs through the release of tumor cells into the blood circulation (circulating tumor cells, CTCs). There has been considerable interest in analyzing CTCs as a potential source of clinically actionable information relating to molecular profile of the patient’s disease. CTCs can be accessed repeatedly and noninvasively and, thus, provide a clinical feasible methodology for tracking longitudinal changes in disease profile that is not readily accomplished with conventional biopsy. In fact, this approach provides only a snapshot of the tumor restricted to the site and state of the cancer at the time of biopsy [2].
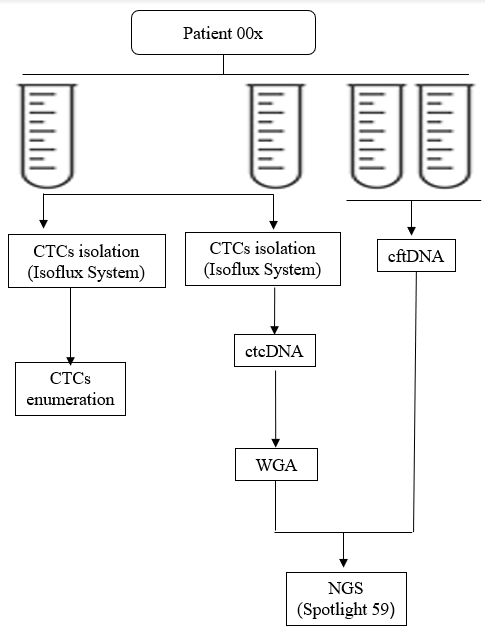
CTCs can enter and stay in the bloodstream long before tumor metastasis occurs and even at very early stage of cancer development [3, 4]. Hence, CTCs not only offer the potential to study the mechanism of cancer metastasis, but also allow ongoing monitoring of cancer progression and prediction of therapy response in a minimally-invasive way, which is referred to as liquid biopsy [5, 6]. To date, a number of studies have investigated the diagnostic and prognostic role of liquid biopsy, mainly through CTCs enumeration [5, 7]. In BC, CTCs represent a promising tool for identifying patients with high risk for tumor relapse in both metastatic and primary BC [8-12]. However, since the tumor is a “moving target”, with significant molecular alterations evolving during progression and in response to therapeutics, extending liquid biopsy to the genetic analysis of CTCs will further help to inform on these two relevant aspects of carcinogenesis and, consequently, to reduce mortality. In addition to CTCs, tumors can release DNA into the bloodstream, which is described as circulating tumor DNA (ctDNA) [13]. This cell-free DNA (cfDNA), although present also in healthy individuals, is significantly increased in cancer subjects [14]. Therefore, ctDNA is recognized as a novel biomarker in the diagnosis of several cancers, including gastric cancer, non-small lung cancer, and hepatocellular carcinoma [15-17]. In BC, the concentration of ctDNA has potential first diagnostic value for BC [13, 18]. Given that the combination of as many parameters as possible may lead to achieve the maximum level of diagnostic and/or prognostic accuracy, in the present study, we carried out liquid biopsy through CTCs enumeration, ctcDNA and cftDNA quantification and sequencing, in early BC patients before and after surgery (Figure 1), providing, for the first time to our knowledge, a complete picture, even though still limited to a small cohort of patients, of this potentially important diagnostic and prognostic strategy.
Methods
I Patients
This is a prospective trial evaluating the liquid biopsy on women with operable BC admitted to the Breast Unit of the Santa Maria della Misericordia Hospital, Perugia, Italy. The study was reviewed and approved by the institution's Ethics Committee (Authorization CEAS Umbria #3068) and conducted in accordance with Good Clinical Practice Guidelines and the Declaration of Helsinki. Informed consent was obtained from all patients. Enrollment was strictly voluntary and patient results were blinded from investigators by use of a random number system as the unique patient identifier. Patients with metastatic disease and with other previous tumors or who underwent neo-adjuvant therapy were excluded from this study. Tumor stage was classified according to the American Joint Commission on Cancer (AJCC) tumor-node-metastasis (TNM) classification and histopathological grading was assessed according to the modified Bloom–Richardson system [19]. Estrogen receptors (ERs) and progesterone receptors (PgRs) were assessed by immunohistochemistry (IHC) and were considered positive if at least 10% of the cells were stained. Tumors were considered HER2 positive if they were scored 3+ by IHC or fluorescence in situ hybridization (FISH) amplified.
II Blood Samples Collection
Four whole peripheral blood (WPB) samples were collected from each patient before and after surgery. Two BD Vacutainer® K2 EDTA Tubes each of 7,5 ml of WPB were used for CTCs isolation and two PAXgene Blood DNA Tubes each of 10 ml WPB were used for cell-free tumor DNA (cftDNA) extraction (Figure 2).
Figure 1: Flow diagram of the study protocol
Figure 2: Flow diagram of the sampling and blood withdraw processing.
III CTCs Isolation
Two EDTA tubes were processed, within 30 min from the initial blood draw, for CTCs isolation. CTCs isolated from one blood tube underwent subsequent CTCs enumeration and those isolated from the second one underwent subsequent lysis, whole genome amplification (WGA) and NGS analysis. CTCs isolation was performed by using the specific CTC Enrichment Kit (Fluxion Biosciences Inc, San Francisco, CA, USA), according to manufacturer’s instructions. Briefly, the peripheral blood mononuclear cell fraction was recovered by using LeucoSep tubes (Greiner Bio-One, Monroe, NC, USA) pre-treated with 15 ml of Ficoll-Paque Plus (GE Healthcare, Pittsburgh, PA, USA) and resuspended in 1 ml of binding buffer. Immunomagnetic beads pre-conjugated with anti-EpCAM antibodies (CTC Enrichment Kit; Fluxion Biosciences Inc) were added directly to the sample and incubated for 2 hours at 4°C with passive mixing on a rotator. Following the magnetic bead coupling step, samples were loaded into the inlet well of the microfluidic cartridge in Isoflux system (Fluxion Biosciences Inc., San Francisco, CA, USA). Samples were flowed through the channel at a flow rate of 20 μl per minute by applying a head pressure of 2 psi to the inlet well in. Each sample passed through the channel in less than 45 minutes. After processing, the disk containing the cells in a hanging drop is inserted into a holder and the isolated target cells were recovered off the isolation zone disk through pipetting and dispensed into a microfuge tube for further processing [3].
IV CTCs Enumeration Analysis
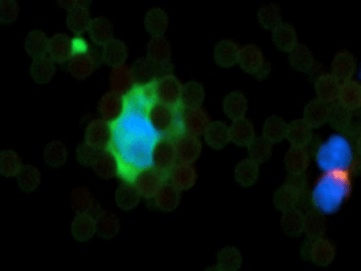
For enumeration, IsoFlux isolated target cells were recovered from the disk through pipetting and immunofluorescence staining was performed using anti-cytokeratin (CK), anti-CD45, and Hoechst 33342 (nucleus) by using IsoFlux CTC Enumeration Kit (Fluxion Biosciences Inc.) and following manufacturer’s instructions. Briefly, recovered CTCs were fixed with phosphate-buffered saline (PBS) buffer containing 1.8% formaldehyde, washed, and blocked with 1% donkey serum in PBS. Cells were stained with rabbit polyclonal anti-human CD45 antibody followed by goat anti-rabbit antibody conjugated with Cy3. After permeabilization with 0.1% Triton X-100, cells were stained with anti-CK (fluorescein isothiocyanate). For CK staining, we used antibody clone CK3-6H5, a pancytokeratin-specific antibody likely to recognize all simple epithelium CKs. Stained CTCs were mounted in SlowFade Gold mounting media with Hoechst 33342 (Life Technologies Inc, Foster City, CA, USA) to Sensoplate Glass-Bottom Multiwell Plates (Greiner Bio-One) for imaging. Cells were scored as CTCs if they were CK+, CD45−, nucleated, and morphologically intact (Figure 3) [3]. Imaging and counting were performed using Cytation Imaging Reader (BioTek, Canada).
V ctcDNA Extraction and Whole Genome Amplification (WGA)
By using the IsoFlux NGS DNA Kit (Fluxion Biosciences Inc.), CTCs were lysed to obtain ctcDNA that was subsequently amplified by whole genome amplification (WGA) reaction, thus generating DNA samples amenable to NGS, according to manufacturer’s instructions [4]. Subsequently the amplified ctcDNA was purified using QIAamp DNA Mini Kit (Qiagen S.p.A., Milan, Italy) and stored at -80°C until its use.
VI cftDNA Extraction
Two PAXgene Blood DNA Tubes, each of 10 ml of WPB, were collected from each patient and processed within 30 min from the initial blood draw for cftDNA extraction. Whole blood was first centrifuged at 1900g for 15 min to separate the plasma from the peripheral blood cells. The supernatant was collected and transferred into a 2 mL Eppendorf tube (EP tube), followed by centrifugation at 1900 g for 10 min to pellet any remaining cells. Plasma (supernatant) was collected, transferred into a new 2 mL EP tube and stored at -80°C. The cftDNA was extracted from the plasma samples (at least 4 mL) using QIAamp MinElute ccfDNA Midi Kit using QIAcube instrument (Qiagen, S.p.A., Milan, Italy), according to the manufacturer’s instruction. The cftDNA was recovered in 55 μl of elution buffer and was stored at -20°C until its use.
Figure 3: A circulating tumor cell (CTC). Cells are scored as CTCs if they are CK+ (green color), CD45- (pink color), nucleated (blue color), morphologically intact and vital. On the right side corner is evident a lymphocyte (CD45+) and nucleated (blue color). Magnification 40x. All around the CTC are shown capturing magnetic beads.
Figure 4: Genes represented in the Spotlight 59 Oncology Panel and Number of Amplicons.
VII ctcDNA and cftDNA by NGS
For NGS analysis of both ctcDNA and cftDNA we used the Spotlight 59 Panels (Fluxion Biosciences Inc, San Francisco, CA, USA), an oncology targeted amplification panel that includes 59 clinically-relevant oncology-related genes (Figure 4) [5]. For the preparation of high quality targeted NGS library, q-PCR on ABI 7300 Real-Time PCR System (Applied Biosystems) was performed using iTaq Universal SYBR Green Supermix and Alu115 and Alu247 primer pairs for an accurate quantification and quality evaluation of the starting DNA. Subsequent step is targeted amplification utilizing the Spotlight 59 kit, a single tube amplification reaction repeated over 2 technical replicates, each starting with a DNA input of 10 ng. The kit includes sample barcoding and provides the reagents required for full preparation of multiplex libraries. Accurate quantitation of libraries by Q-PCR was performed using NEBNext® Library Quant Kit (BioLabs Inc.) to maximize data output and quality from each sequencing run. Samples were then sequenced on MiSeq Illumina instrument to a mean depth of 5,000x per replicate, 10,000x per sample. Molecular profile analysis was determined using ERASE-Seq (Elimination of Recurring Artifacts and Stochastic Errors), a method for the accurate detection and sensitivity of low frequency DNA ranges. The Spotlight 59 panel and the ERASE-Seq workflow determine a sensitivity > 90% and < 0.1 false positives at 0.1% of the allele frequency [6].
VIII Statistical Analysis
T-test, Cochran-Armitage test for trend or χ-square (without unknowns, if applicable), the statistical evaluation we employ, assesses the associations between the presence of CTC and the primary characteristics of the tumor. P values were two-tailed and considered significant if P < 0.05.
Table 1: Presence of CTCs at baseline according to clinical-pathological variables.
|
|
All patients |
Patients without CTCs |
Patients with CTCs |
P-value |
|
|
(n = 47) |
(n = 9 )* |
(n = 36 )* |
|
|
|
n (%) |
n (%) |
n (%) |
|
|
Age (years) |
|
|
|
|
|
Mean ± SD |
54.4 ± 9.8 |
57 ± 8.2 |
53.8 ± 10.1 |
0.314a |
|
Range |
33-71 |
47-69 |
33-71 |
|
|
Tumor stage |
|
|
|
|
|
pT1 |
34 (72.3) |
7 (77.8) |
27 (75.0) |
0.730b |
|
pT2 |
7 (14.9) |
1 (11.1) |
5 (13.8) |
|
|
pT3 |
2 (4.3) |
0 |
2 (5.6) |
|
|
pT4 |
1 (2.1) |
- |
- |
|
|
Unknown |
3 (6.4) |
1 (11.1) |
2 (5.6) |
|
|
Nodal stage |
|
|
|
|
|
pN0 |
30 (63.8) |
4 (44.5) |
25 (69.4) |
0.163b |
|
pN1 |
7 (14.9) |
1 (11.1) |
6 (16.7) |
|
|
pN2 |
3 (6.4) |
0 |
2 (5.6) |
|
|
pN3 |
3 (6.4) |
2 (22.2) |
1 (2.7) |
|
|
Unknown |
4 (8.5) |
2 (22.2) |
2 (5.6) |
|
|
Histological grading |
|
|
|
|
|
G1 |
13 (27.7) |
3 (33.3) |
10 (27.7) |
0.738b |
|
G2 |
25 (53.2) |
5 (55.6) |
18 (50) |
|
|
G3 |
7 (14.9) |
1 (11.1) |
6 (16.7) |
|
|
unknown |
2 (4.2) |
0 |
2 (5.6) |
|
|
Histological type |
|
|
|
|
|
Ductal |
39 (83) |
5 (55.6) |
32 (88.8) |
0.562b |
|
Lobular |
5 (10.6) |
3 (33.3) |
2 (5.6) |
|
|
In situ |
3 (6.4) |
1 (11.1) |
2 (5.6) |
|
|
Hormone receptor status (ER, PgR) |
|
|
|
|
|
Negative |
5 (10.7) |
0 |
5 (13.8) |
0.299c |
|
Positive |
41 (87.2) |
9 (100) |
30 (83.5) |
|
|
Unknown |
1 (2.1) |
0 |
1 (2.7) |
|
|
Ki67** |
|
|
|
|
|
High |
13 (27.7) |
2 (22.2) |
10 (27.8) |
0.569c |
|
Low |
30 (63.8) |
6 (66.7) |
23 ( 63.9) |
|
|
Unknown |
4 (8.5) |
1 (11.1) |
3 (8.3) |
|
|
HER2 status |
|
|
|
|
|
Negative |
38 (80.9) |
6 (66.7) |
30 (83.4) |
0.246c |
|
Positive |
5 (10.6) |
2 (22.2) |
3 (8.3) |
|
|
Unknown |
4 (8.5) |
1 (11.1) |
3 (8.3) |
|
|
Type of surgery |
|
|
|
|
|
Breast conserving |
34 (72.4) |
7 (77.8) |
26 (72.2) |
0.550c |
|
Mastectomy |
13 (27.6) |
2 (22.2) |
10 (27.8) |
|
at-test
bCochran-Armitage test for trend (without unknowns when appropriate)
cχ2 test (without unknowns when appropriate)
* Total CTCs n = 45 (for two patients no CTCs samples were available) **ki67 was considered high (> 20%)
Results
I Patient Characteristics and Circulating Tumor Cell (CTCs) status
In this explorative study we reported the preliminary results of 50 patients enrolled at the Breast Unit of the Santa Maria della Misericordia Hospital in Perugia from April 2018 to February 2019. Of the 50 patients included in the study, 44 patients had infiltrative BC, 3 patients had in situ BC and 3 patients were healthy controls (Figure 1). Patient characteristics are described in (Table 1). The median diagnostic age of the 47 patients was 54 years (range 33-71 years) and all patients were female. The majority of patients (93.6%) had invasive carcinoma and the 87.2% had ER/PR positive disease. There were no statistically significant associations between the number of CTCs and clinicopathological data (Table 1). In total, 45 patients with known CTCs status at baseline (for two pts CTCs status were not available) were included in the final analysis. One or more CTCs were detected in 80% (36 out of 45) of samples. All healthy controls were CTCs-negative.
Among the three in situ BC patients two presented 1 and 3 CTCs, while 1 had 0 CTCs. For only one patient, the number of CTC after surgery was available and showed the same number of CTCs before the treatment (n = 3). Among the infiltrative BC group, 44 patients provided a pre-surgery and 21 post-surgery sample. Hence, we enumerated CTCs in a total of 21 patients before and after surgical therapy. By comparing, in this group, the number of CTCs before and after surgery, we noticed that 66% of patients who presented CTCs before surgery showed a decreased number of CTCs after surgery (Table 2, Row A.). Conversely, 14% of patients who were positive for CTCs before surgery, continued to have the same number of CTCs after surgery (Table 2, Row B.). Interestingly, 19% of patients who had a pre-surgery low number of CTCs showed an increased number of CTCs post-surgery (Table 2, Row C.). Total cftDNA was obtained for 22 samples. Levels of circulating tumor DNA varied; median concentration was 23 ng/µl (range 0.01-0.49 ng/µl). The mean plasma mutant allele frequency (AF) ranged from 0.22% to 2.01%.
II Mutation profiling of ctcDNA and cftDNA by NGS
We assessed by NGS the mutation profiling of ctcDNA and matching cftDNA in samples of patients taken prior surgery. Although we examined mutation hotspots in 59 genes by NGS, the preliminary results showed that 52% (11 out of 21) of ctcDNA samples analyzed had mutations in 9 genes (TP53, CDKN2A, FBXW7, PTPN11, KRAS, NRAS, BRAF, IDH1, ALK) and only one mutation in one gene was detected in the majority of samples. The patient 0022 had four mutations in four genes (ALK, KRAS, NRAS, CDKN2A). TP53 and ALK were the most frequently mutated genes. In the patient 0004, sequencing of ctcDNA revealed the presence of two mutations in TP53 gene (p.R282W, p.A161T) and 1 mutation in ALK gene (p.F1174C). Two patients who had 0 CTCs were sequenced as control and no mutations were found. The cftDNA was sequenced only for six patients and of them, three patients had mutations in 5 genes (PIK3CA, APC ALK, KRAS, TSC1) whereas in the remaining three patients no mutations were detected. In one patient (0015) who did not present mutations in the ctcDNA (CTC = 0) we determined 1 mutation in the APC gene (p.K1308*) from cftDNA plasma (Table 3).
Table 2: Number of circulating tumor cells (CTCs) pre- and post-surgery.
|
A. Patients with CTCs number decreased after surgery |
||
|
|
Number of CTCs/7.5 ml blood |
|
|
Patient ID |
Pre-surgery (A)* |
Post-surgery (B)** |
|
0008 |
28 |
0 |
|
0013 |
10 |
1 |
|
0014 |
7 |
0 |
|
0044 |
6 |
0 |
|
0005 |
6 |
1 |
|
0037 |
4 |
0 |
|
0006 |
7 |
4 |
|
0029 |
7 |
3 |
|
0026 |
3 |
2 |
|
0036 |
5 |
3 |
|
0004 |
2 |
0 |
|
0020 |
1 |
0 |
|
0042 |
3 |
1 |
|
0049 |
7 |
2 |
|
B. Patients with CTCs number persisted after surgery |
||
|
0003 |
1 |
1 |
|
0017 |
3 |
3 |
|
0018 |
3 |
3 |
|
C. Patients with CTCs number increased after surgery |
||
|
0016 |
1 |
2 |
|
0019 |
0 |
1 |
|
0022 |
2 |
4 |
|
0030 |
0 |
4 |
*(A) sample collected pre-surgery; **(B) sample collected post-surgery
Discussion
Cancer therapy decisions are often made according to the histopathological-molecular profile of tumor tissue obtained from surgery or biopsy. It has been shown that tumor profiles change with time and treatment, and that tumor tissue is heterogeneous. Thus, other approaches that are easily accessible and less invasive than surgery or biopsy to monitor responses to treatment and predict relapses are urgently needed. In the last few years, the term "liquid biopsy" (LB) has been introduced to represent multifunctional circulating biomarkers in the peripheral blood and other physiological fluids of patients with cancer. LB is a noninvasive alternative to tissue biopsies, but it has not been implemented in routine clinical practice for BC. In addition, liquid biopsy seems to be a promising approach for personalized medicine, which enables the prediction, monitoring, and rational selection of appropriate therapy for individual patients.
Although unequivocal evidence has shown the prognostic relevance of CTCs in the peripheral blood of patients with metastatic BC, less evidence is available for the prognostic relevance of CTCs at the time of primary diagnosis. In the present study, we conducted LB through CTCs enumeration, ctcDNA and cftDNA quantification and sequencing for mutational analysis in early BC patients before and after surgery (Figure 1), providing, for the first time to our knowledge, the characterization, even though still limited to a small cohort of patients, of several prognostic tumor biomarkers, mirroring BC phenotype especially in its evolution. Specifically, we found that surgery was able to decrease the number of CTCs in a significant percentage of patients (66%). On a biological point of view, this is consistent with the assumption that cancer feed the release of malignant cells into the bloodstream, hence the reduction or reset of CTCs mirrors the removal of the tumor source. Dissemination of tumor cells from the primary tumor into the bloodstream is a critical step in tumorigenesis and is considered a precursor of distant metastases [12]. CTCs have been defined “silent predictors of metastases”, this means that CTCs identifies “in progress” metastases onset, even before any other high-resolution imaging method is able to do [20].
Hence, our results would suggest a potential resolution of the disease for those patients with no CTCs after surgery. Conversely, in 33% of patients the number of CTCs either remained unchanged or increased after surgical therapy. Hence, if the number of CTCs after surgery does not reset, the presence of distant micro-metastatic foci can be suspected that supply the circulatory torrent, with consequent persistence of the residual disease after surgery. In all cases, the enrolling of additional patients (which is ongoing in our laboratory) and the follow-up of those already collected are warranted, in order to prove our preliminary hypotheses from this explorative study. As above mentioned, isolation and characterization of the molecular profile of CTCs has the potential to provide a far easier liquid biopsy than tumor tissue biopsies to monitor malignant cell populations during disease progression and in response to therapies. Several CTCs capture and/or isolation technologies have been developed. The bead-based EpCAM antibody CTC capturing system is most widely used and one of them, Cell Search, the only Food and Drug Administration (FDA) approved device for CTCs analysis (enumeration), is considered the current “gold standard” [21].
Table 3: ctcDNA and cftDNA mutations identified by NGS.

*AF: Allele Frequency
However, the performance and applicability of this system is receiving more and more challenge, especially regarding the capture rate (low cell recovery) and purity (low purity). Moreover, a key limitation of this technology is the ability to release and recover viable intact CTCs for downstream molecular analysis. Here, we successfully detected, counted and lysed CTCs (to get ctcDNA for subsequent NGS) from patients with early BC using the Isolation System Isoflux (Fluxion, San Francisco, CA). Although also Isoflux is an immunobead-based system, it has been projected to address Cell Search limitations, by providing high-sensitivity rare cell isolation coupled with a novel cell retrieval microfluidic-based mechanism [2]. We, here, confirm that Isoflux provides a complete workflow to track oncogene mutational changes longitudinally with high success rates. Regarding the analysis of the mutation profiling of ctcDNA and matching cftDNA, our preliminary results refers to early-stage BC samples taken prior to surgery. This was a challenging issue of the study, since, as known, the fraction of ctcDNA and the total concentration of cftDNA are directly correlated with the stage of the disease and are significantly lower in earlier stages, which posed some technical issues in the extraction process and subsequent mutational analysis [22].
Although we examined mutation hotspots in 59 genes by NGS, in the sequenced ctcDNA, the mutations were only detected in 9 genes (TP53, CDKN2A, FBXW7, PTPN11, KRAS, NRAS, BRAF, IDH1, ALK) and in the sequenced cftDNA, in 5 genes (PIK3CA, APC ALK, KRAS, TSC1). TP53 was the most frequently mutated gene (Figure 4). On a biological point of view, this is consistent with the assumption that a non-functional TP53 has been shown to offer survival advantages to the cancer cells by eluding apoptosis, facilitating growth, conferring anoikis resistance, and the emergence of a potentially more aggressive malignancy [23]. TP53 mutations are exceptionally frequent in cancer and are among the key driving factors in triple-negative breast cancer (TNBC) [24]. Furthermore, TP53 mutations have been shown to predict a poor response to anthracycline-based neoadjuvant chemotherapy; others suggested that TP53 mutations confer sensitivity to taxane [25-27]. A recent study suggested that patients with TP53 mutations are more likely to respond to anthracycline/cyclophosphamide-based neoadjuvant chemotherapy [28]. Another gene that is frequently mutated in BC patients is PIK3CA. Oshiro et al. reported that mutations in the PI3KCA gene could be targeted as a marker predictive of recurrence [29].
In our study, at present, one patient (0022) had PIK3CA mutated (p. R93W) in the cftDNA. Although promising, this result is still far from being interpreted in the context of the literature due to the small number of patients (n = 6) characterized for their molecular profile by NGS. In addition, in this patient (0022) a different molecular profile of the ctcDNA compared to that of the cftDNA was found. It has been successfully demonstrated by single-cell sequencing that many breast cancers are composed of multiple distinct subclones [30]. Intra-patient cellular heterogeneity is widely reported in epithelial malignancies and it is expected that CTCs will also be heterogeneous [31]. Our results are consistent with previous findings which showed a heterogeneous pattern of genomic mutations on single ctcDNA or cftDNA obtained from several cancer patients, including BC ones.
Conclusions
This study showed that the enumeration of CTCs might be an important tool to monitor the progress of the disease and that the isolation of both ctcDNA and cftDNA from early BC patients can be used for genomic analysis, both to initially identify targetable mutations and to be used as a biomarker to reveal which cell populations are affected by the current or previous therapy. As required by our study protocol, collecting sequential blood samples for real-time monitoring of the efficacy of systemic therapies would offer new possibilities in evaluating targeted therapies based on genomic profiling of ctcDNA and cftDNA, so improving the clinical management of patients with early BC.
Ethics Approval and Consent to Participate
This study was approved by our Institutional Board Comitato Etico delle Aziende Sanitarie della Regione Umbria (CEAS Umbria- Istituito con Delibera di Giunta Regionale del 05/03/2007) Authorization CEAS Umbria #3068 in date September 14th, 2017. All patients provided written informed consent.
Consent for publication
Not applicable.
Conflicts of interest
All authors declare that they have no competing interests.
Funding
This study was supported by the Italian League for the fight against cancer - LILT (Ministero della Salute, 5 x l000 year 2014). The funders only acted as economic support and did not actively participate in the development of the research.
Author Contributions
RA designed the study; RA, LV, AC reviewed and analyzed the clinical data and did the critical revision of the manuscript; SA, LV performed the NGS, analyzed the data and drafted the manuscript; AC performed CTC analyses; TV reviewed the manuscript; ZS data manager; IL reviewed the manuscript; CP enrolled the patients. All authors read and approved the manuscript final version.
Acknowledgments
We thank Roberta Frosini for her technical assistance.
Abbreviations
BC: Breast Cancer
Ctcs: Circulating Tumor Cells
Cftdna: Cell-Free Tumor DNA
NGS: Next Generation Sequencing
ER: Estrogen Receptor
PR: Progesterone Receptor
FISH: Fluorescence in Situ Hybridization
HER2: Human Epidermal Growth Factor Receptor 2
IHC: Immunohistochemistry
Q-PCR: Quantitative Polymerase Chain Reaction
Article Info
Article Type
Research ArticlePublication history
Received: Fri 20, Dec 2019Accepted: Thu 02, Jan 2020
Published: Fri 10, Jan 2020
Copyright
© 2023 Antonio Rulli. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.ACO.2020.01.01
Author Info
Antognelli Cinzia Antonio Rulli Covarelli Piero Izzo Luciano Ludovini Vienna Siggillino Annamaria Talesa Vincenzo Nicola Zayik Svitlana
Corresponding Author
Antonio RulliDepartment of Biomedical and Surgical Sciences, Division of Surgical Oncology, University of Perugia, Perugia, Italy
Figures & Tables
Table 1: Presence of CTCs at baseline according to clinical-pathological variables.
|
|
All patients |
Patients without CTCs |
Patients with CTCs |
P-value |
|
|
(n = 47) |
(n = 9 )* |
(n = 36 )* |
|
|
|
n (%) |
n (%) |
n (%) |
|
|
Age (years) |
|
|
|
|
|
Mean ± SD |
54.4 ± 9.8 |
57 ± 8.2 |
53.8 ± 10.1 |
0.314a |
|
Range |
33-71 |
47-69 |
33-71 |
|
|
Tumor stage |
|
|
|
|
|
pT1 |
34 (72.3) |
7 (77.8) |
27 (75.0) |
0.730b |
|
pT2 |
7 (14.9) |
1 (11.1) |
5 (13.8) |
|
|
pT3 |
2 (4.3) |
0 |
2 (5.6) |
|
|
pT4 |
1 (2.1) |
- |
- |
|
|
Unknown |
3 (6.4) |
1 (11.1) |
2 (5.6) |
|
|
Nodal stage |
|
|
|
|
|
pN0 |
30 (63.8) |
4 (44.5) |
25 (69.4) |
0.163b |
|
pN1 |
7 (14.9) |
1 (11.1) |
6 (16.7) |
|
|
pN2 |
3 (6.4) |
0 |
2 (5.6) |
|
|
pN3 |
3 (6.4) |
2 (22.2) |
1 (2.7) |
|
|
Unknown |
4 (8.5) |
2 (22.2) |
2 (5.6) |
|
|
Histological grading |
|
|
|
|
|
G1 |
13 (27.7) |
3 (33.3) |
10 (27.7) |
0.738b |
|
G2 |
25 (53.2) |
5 (55.6) |
18 (50) |
|
|
G3 |
7 (14.9) |
1 (11.1) |
6 (16.7) |
|
|
unknown |
2 (4.2) |
0 |
2 (5.6) |
|
|
Histological type |
|
|
|
|
|
Ductal |
39 (83) |
5 (55.6) |
32 (88.8) |
0.562b |
|
Lobular |
5 (10.6) |
3 (33.3) |
2 (5.6) |
|
|
In situ |
3 (6.4) |
1 (11.1) |
2 (5.6) |
|
|
Hormone receptor status (ER, PgR) |
|
|
|
|
|
Negative |
5 (10.7) |
0 |
5 (13.8) |
0.299c |
|
Positive |
41 (87.2) |
9 (100) |
30 (83.5) |
|
|
Unknown |
1 (2.1) |
0 |
1 (2.7) |
|
|
Ki67** |
|
|
|
|
|
High |
13 (27.7) |
2 (22.2) |
10 (27.8) |
0.569c |
|
Low |
30 (63.8) |
6 (66.7) |
23 ( 63.9) |
|
|
Unknown |
4 (8.5) |
1 (11.1) |
3 (8.3) |
|
|
HER2 status |
|
|
|
|
|
Negative |
38 (80.9) |
6 (66.7) |
30 (83.4) |
0.246c |
|
Positive |
5 (10.6) |
2 (22.2) |
3 (8.3) |
|
|
Unknown |
4 (8.5) |
1 (11.1) |
3 (8.3) |
|
|
Type of surgery |
|
|
|
|
|
Breast conserving |
34 (72.4) |
7 (77.8) |
26 (72.2) |
0.550c |
|
Mastectomy |
13 (27.6) |
2 (22.2) |
10 (27.8) |
|
at-test
bCochran-Armitage test for trend (without unknowns when appropriate)
cχ2 test (without unknowns when appropriate)
* Total CTCs n = 45 (for two patients no CTCs samples were available) **ki67 was considered high (> 20%)
Table 2: Number of circulating tumor cells (CTCs) pre- and post-surgery.
|
A. Patients with CTCs number decreased after surgery |
||
|
|
Number of CTCs/7.5 ml blood |
|
|
Patient ID |
Pre-surgery (A)* |
Post-surgery (B)** |
|
0008 |
28 |
0 |
|
0013 |
10 |
1 |
|
0014 |
7 |
0 |
|
0044 |
6 |
0 |
|
0005 |
6 |
1 |
|
0037 |
4 |
0 |
|
0006 |
7 |
4 |
|
0029 |
7 |
3 |
|
0026 |
3 |
2 |
|
0036 |
5 |
3 |
|
0004 |
2 |
0 |
|
0020 |
1 |
0 |
|
0042 |
3 |
1 |
|
0049 |
7 |
2 |
|
B. Patients with CTCs number persisted after surgery |
||
|
0003 |
1 |
1 |
|
0017 |
3 |
3 |
|
0018 |
3 |
3 |
|
C. Patients with CTCs number increased after surgery |
||
|
0016 |
1 |
2 |
|
0019 |
0 |
1 |
|
0022 |
2 |
4 |
|
0030 |
0 |
4 |
*(A) sample collected pre-surgery; **(B) sample collected post-surgery
Table 3: ctcDNA and cftDNA mutations identified by NGS.

*AF: Allele Frequency

References
- I NUMERI DEL CANCRO IN ITALIA 2017.
- Harb W, Fan A, Tran T, Danila DC, Keys D et al. ( 2013) Mutational Analysis of Circulating Tumor Cells Using a Novel Microfluidic Collection Device and qPCR Assay. Transl Oncol 6: 528-538. [Crossref]
- Hüsemann Y, Geigl JB, Schubert F, Musiani P, Meyer M et al. (2008) Systemic Spread Is an Early Step in Breast Cancer. Cancer Cell 2008 13: 58-68. [Crossref]
- Eyles J, Puaux A-L, Wang X, Toh B, Prakash C et al. (2010) Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J Clin Invest 120: 2030-2039. [Crossref]
- Xu L, Mao X, Imrali A, Syed F, Mutsvangwa K et al. (2015) Optimization and Evaluation of a Novel Size Based Circulating Tumor Cell Isolation System. PLoS One 10: e0138032. [Crossref]
- Lu Y-J, Xu L, Shamash J (2015) Circulating Tumor Cells: A Window to Understand Cancer Metastasis, Monitor and Fight Against Cancers. J Can Res Updates 4: 13-29.
- Parkinson DR, Dracopoli N, Petty BG, Compton C, Cristofanilli M et al. (2012) Considerations in the development of circulating tumor cell technology for clinical use. J Transl Med 10: 138. [Crossref]
- Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J et al. (2004) Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N Engl J Med 351: 781-791. [Crossref]
- Franken B, de Groot MR, Mastboom WJ, Vermes I, van der Palen J et al. (2012) Circulating tumor cells, disease recurrence and survival in newly diagnosed breast cancer. Breast Cancer Res 14: R133. [Crossref]
- Rack B, Schindlbeck C, Jückstock J, Andergassen U, Hepp P et al. (2014) Circulating Tumor Cells Predict Survival in Early Average-to-High Risk Breast Cancer Patients. J Natl Cancer Inst 106. [Crossref]
- Akbari MR, Lepage P, Rosen B, Mc Loughlin J, Risch H et al. (2014). J Natl Cancer Inst 106.
- Janni WJ, Rack B, Terstappen LW, Pierga JY, Taran FA et al. (2016) Pooled Analysis of the Prognostic Relevance of Circulating Tumor Cells in Primary Breast Cancer. Clin Cancer Res 22: 2583-2593. [Crossref]
- Yu D, Tong Y, Guo X, Feng L, Jiang Z et al. (2019) Diagnostic Value of Concentration of Circulating Cell-Free DNA in Breast Cancer: A Meta-Analysis. Front Oncol 9: 95. [Crossref]
- Mouliere F, El Messaoudi S, Pang D, Dritschilo A, Thierry AR (2014) Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol Oncol 8: 927-941. [Crossref]
- Gao Y, Zhang K, Xi H, Cai A, Wu X et al. (2017) Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: a meta-analysis. Oncotarget 8: 6330-6340. [Crossref]
- Jiang T, Zhai C, Su C, Ren S, Zhou C (2016) The diagnostic value of circulating cell free DNA quantification in non-small cell lung cancer: A systematic review with meta-analysis. Lung Cancer 100: 63-70. [Crossref]
- Liao W, Mao Y, Ge P, Yang H, Xu H et al. (2015) Value of quantitative and qualitative analyses of circulating cell-free DNA as diagnostic tools for hepatocellular carcinoma: a meta-analysis. Medicine (Baltimore) 94: e722. [Crossref]
- Wang R, Li X, Zhang H, Wang K, He J (2017) Cell-free circulating tumor DNA analysis for breast cancer and its clinical utilization as a biomarker. Oncotarget 8: 75742-75755. [Crossref]
- Singletary SE, Allred C, Ashley P, Bassett LW, Berry D et al. (2002) Revision of the American Joint Committee on Cancer Staging System for Breast Cancer. J Clin Oncol 20: 3628-3636. [Crossref]
- Pantel K, Alix-Panabières C, Riethdorf S (2009) Cancer micrometastases. Nat Rev Clin Oncol 6: 339-351. [Crossref]
- Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC et al. (2004) Tumor Cells Circulate in the Peripheral Blood of All Major Carcinomas but not in Healthy Subjects or Patients With Nonmalignant Diseases. Clin Cancer Res 10: 6897-6904. [Crossref]
- Chen KZ, Lou F, Yang F, Zhang JB, Ye H et al. (2016) Circulating Tumor DNA Detection in Early-Stage Non-Small Cell Lung Cancer Patients by Targeted Sequencing. Sci Rep 6: 31985. [Crossref]
- Zhang Y, Lu H, Dazin P, Kapila Y (2004) Squamous Cell Carcinoma Cell Aggregates Escape Suspension-induced, p53-mediated Anoikis. J Biol Chem 279: 48342-48349. [Crossref]
- Walerych D, Napoli M, Collavin L, Del Sal G (2012) The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis 33: 2007-2017. [Crossref]
- Berns EM, Foekens JA, Vossen R, Look MP, Devilee P et al. (2000) Complete sequencing of TP53 predicts poor response to systemic therapy of advanced breast cancer. Cancer Res 60: 2155-2162. [Crossref]
- Kandioler-Eckersberger D, Ludwig C, Rudas M, Kappel S, Janschek E et al.(2000) TP53 mutation and p53 overexpression for prediction of response to neoadjuvant treatment in breast cancer patients. Clin Cancer Res 6: 50-56. [Crossref]
- Glück S, Ross JS, Royce M, McKenna EF, Perou CM et al. (2012) TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer Res Treat 132: 781-791. [Crossref]
- Wang Y, Xu Y, Chen J, Ouyang T, Li J et al. (2016) TP53 mutations are associated with higher rates of pathologic complete response to anthracycline/cyclophosphamide-based neoadjuvant chemotherapy in operable primary breast cancer. Int J cancer 138: 489-496. [Crossref]
- Oshiro C, Kagara N, Naoi Y, Shimoda M, Shimomura A et al. (2015) PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res Treat 150: 299-307. [Crossref]
- Robinson DR, Wu Y-M, Vats P, Su F, Lonigro RJ et al. (2013) Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet 45: 1446-1451. [Crossref]
- Marusyk A, Almendro V, Polyak K (2012) Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 12: 323-334. [Crossref]
