Microbiome in GIT Cancers: New Frontiers in Personalized Medicine – A Review of Literature
A B S T R A C T
Every person has a characteristic “enterotype,” the gut components and the environment. Recently, a strong biological correlation has emerged among the microbiome of the gut, the immune system, cancer development and pharmacological effects of chemotherapy. In this review, we outline the role of gut microbiota in the gastrointestinal tract (GIT) cancers pathogenesis and their implications for enhancing the efficacy of GIT cancer management in clinical practice. We also summarize the molecular pathways linking gut microbiota and GIT cancers and the effectiveness of manipulating microbiota in GIT cancer therapies such as personalized cancer therapy.
Keywords
Gut microbiome, gastrointestinal tract, cancer, therapy, molecular pathogenesis
Introduction
Gut microbiome is the total amount of microorganisms, bacteria, viruses, protozoa, fungi, and their totality of genetic material found in the gastrointestinal tract (GIT). The microbiome help digesting the food, regulate, train our immune system, protect against other bacteria that cause disease, and produce vitamins including B vitamins (B12, thiamine and riboflavin) and Vitamin K, which is important for blood coagulation [1]. The gut microbiota has a vital role in the absorption of minerals and nutrients, the production of amino acids and enzymes, and the synthesis of short-chain fatty acids (SCFAs). Propionate, butyrate, and acetate are important for gut health, promote the integrity of the epithelial barrier, and yield immunomodulation and protection against pathogens and supply energy for epithelial cells. These are the byproducts of the fermentation [2].
Autoimmune diseases such as diabetes, rheumatoid arthritis, muscular dystrophy, multiple sclerosis, and fibromyalgia are strongly related to microbiome dysfunction. Disease-causing microbes' accumulation over time causes changing gene activity and metabolic processes and that result in an abnormal immune response against substances and tissues present in the body. Autoimmune diseases appear to be passed in families by inheriting the family's microbiome not by DNA inheritance [3, 4].
Gut microbiota in humans is divided into different groups each is called phyla. It consists basically of four major phyla including Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria [5]. However, bacteria settle down in our body, they live in the GIT, skin, oral cavity, vagina, and placenta, most of the bacteria inhabit the GIT, with most of the anaerobic bacteria inhabit the colon. Our bodies have 20,000 eukaryotic genes whereas the gut microbiota has 3.3 million prokaryotic genes [6].
Dysbiosis is the modulation in microbial set resulting in decreased variation and count of commensal bacteria. Some studies suppose that there are relations between gut dysbiosis and chronic diseases such as metabolic syndrome, obesity, inflammatory bowel diseases, cardiovascular diseases, and cancer [7]. Homeostasis between the microbiome and host is maintained by the multilayer intestinal barrier. Disturbance of the balanced relationship might lead to an impaired intestinal barrier, chronic inflammation, and dysbiosis, which are key factors in carcinogenesis [8].
Cancer is a prime global health issue causing over eight million deaths worldwide. It is a compound, multifactorial disease involving mutations in the genetic material, which is regulated by host and environmental interplay. According to the latest global cancer statistics in 2018, colorectal cancer, gastric cancer, and esophageal cancer are the third, fifth and seventh most common cancer types worldwide, respectively. Colorectal cancer, gastric cancer, and esophageal cancer are the second, third and sixth leading causes of cancer-related deaths, respectively. Understanding of the direct and indirect relationship between microbiome and GIT cancers can offer new insights for cancer prevention and treatment [9].
Numerous studies have elucidated that the carcinogenicity is mainly attributed to microbial dysbiosis like chronic inflammation with DNA damage in the epithelium, aberrant DNA methylation, increased interleukin, and tumor necrosis factor-α (TNF-α) levels with the activation of mitogen-activated protein kinases (MAPK) pathway, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), and Wnt signaling and, the inhibition of apoptosis, and increase oxidative stress, thus activating proto-oncogenes (e.g., KRAS mutation) also inactivate tumor-suppressor genes (e.g., P53 mutation), triggers a number of innate immune responses involved in the tumor formation process by recognizing the structural components of bacteria, such as lipopolysaccharide (LPS), and flagellin through Toll-like receptors (TLRs) and Nod-like receptors (NLRs) modulating gut microbial composition and enhancing dysbiosis mediated by the inflammasome [10-13]. Also, B cells, T helper (Th) cells, and T regulatory (Treg) cells participate in tumorigenesis through the adaptive immune system, of note, microbial metabolites like lipoteichoic acid, and short-chain fatty acids (SCFAs) have critical roles in GIT cancer through binding to TLR2 [14, 15]. These aforementioned mechanisms may contribute to GIT malignant transformation.
In this review of literature, we have tried to highlight the role of microbiota in the pathogenesis of GIT cancers. Moreover, we have addressed the scientists’ efforts to uncover the mystery of oncomicrobiomes as a promising strategy in GIT cancer diagnosis and treatment. We have searched using PubMed and google scholar databases using the following keywords, microbiome, GIT, cancer, diagnosis, and treatment in the period between 2000 and 2020.
I Role of Microbiome in GIT Cancers Pathogenesis (Table 1)
i Microbiome and Oral Cancer
It is of great importance to determine which bacterial species are responsible for driving oral tumorigenesis. Alterations in the oral commensal microbial sets have potential application in diagnosis, to predict oral squamous cell carcinoma (OSCC ) [16].
The most frequently affected molecular mechanisms by the microbiota to induce carcinogenesis involve epithelial-mesenchymal transition – (EMT) dependent barrier changes and tumor-inducing inflammation [17]. Bacterial infection leads to the emergence of oral cancer and it’s the main cause of chronic inflammation that favours mutagenesis, oncogene activation, cell proliferation, and angiogenesis [18]. It leads to increased expression of matrix metalloproteinase- (MMP-1) and (MMP-10) and activation of (EMT) factors Zeb1 Snail, and Slug without requiring inhibition of miR-200b or E-cadherin [19]. Moreover, it increases the expression of the cancer stem cell markers such as CD133 and CD44 [20]. Many bacterial species are involved in the pathogenesis of oral cancer, Mager et al. reported that Fusobacterium periodonticum was found in the saliva of OSCC patients. Capnocytophaga gingivalis, Prevotella melaninogenica, and Streptococcus mitis were much abundant in saliva of OSCC patients and may serve as a potential marker in the diagnosis of oral cancer [21, 22].
Table 1: Showing summary of microbiome involved in GIT cancers.
|
Disease |
Microbiome biomarker |
Reference |
|
Oral squamous cell carcinoma |
Fusobacterium periodonticum Prevotella melaninogenica Capnocytophaga gingivalis Streptococcus mitis Lactobacillaceae Lactobacillus Haemophilus Aggregatibacter Neisseria Gemellaceae Veillonella Porphyromonas Actinomyces Clostridium Enterobacteriaceae Pseudomonas Atopobium Campylobacter concisus Lautropia mirabilis Rothia dentocariosa Human papilloma virus (HPV) |
[17, 21-27] |
|
Esophageal adenocarcinoma |
Treponemadenticola Streptococcus anginosus Streptococcus mitis Campylobacter Clostridium Lactobacillales Escherichia coli HPV |
[33, 35, 37, 38] |
|
Esophageal squamous cell carcinoma |
Clostridiales Erysipelotrichales Porphyromonas gingivalis Lautropia, Bulleidia, Catonella, Corynebacterium, Moryella, Peptococcus Cardiobacterium Fusobacterium nucleatum |
[40-42, 43] |
|
Gastric cancer |
H. pylori Epstein-Barr virus (EBV) |
[8, 44, 45, 48] |
|
Hepatocellular cancer |
Helicobacter spp. Erysipelotrichaceae Leuconostocaceae Fusobacterium Prevotella Odoribacter Butyricimonas Dorea HBV HCV |
[52, 54, 55]
|
|
Pancreatic Cancer |
Porphyromonas gingivalis, Fusobacterium Neisseria elongata Streptococcus mitis HBV and HCV |
[72, 73] |
|
Colorectal Cancer |
Bacteroides fragilis Enterococcus faecalis Fusobacterium nucleatum Streptococcus gallolyticus Escherichia coli Roseburia Clostridium Faecalibacterium Bifidobacterium Collinsella Peptostreptococcus Prevotella Shigella Ruminococcus Alistipes Veillonella Coprobacter Clostridium symbiosum C. hathewayi |
[75, 76, 78-80] |
Lactobacillus and Lactobacillaceae were significantly abundant in saliva of patients treated with chemo-radiation therapy/ surgery. Patients with advanced TNM stage showed an increased abundance of Lactobacillus in their saliva. Lactobacillus has been stated as the main cause of xerostomia and caries. Haemophilus, Aggregatibacter, Neisseria and Gemellaceae showed higher abundance in healthy individuals when compared to oral cancer patients. Viable bacteria found in deep parts of OSCC supported the hypothesis that bacteria survived in the tumor microenvironment [23, 24]. Microbiome which is of high abundance in tumor sites of OSCC patients: bacterial taxa Veillonella, Porphyromonas, Prevotella, Fusobacterium, Actinomyces, Haemophilus, Enterobacteriaceae, Clostridium belonging anaerobes, and Streptococcus subspecies belonging to aerobes [25, 17]. Atopobium, Pseudomonas, and Capnocytophaga at the genus level and Campylobacter concisus, Prevotellaloeschii, Prevotellasalivae, and Fusobacterium oral taxon 204 at the species level were stated to be significantly abundant in OSCC. Microbiome which plays an important role in inflammation in tumorigenesis are Streptococcus mitis, Streptococcus oral taxon 070, Lautropia mirabilis, and Rothia dentocariosa found in fibroepithelial polyp (FEP) controls with more abundant virulence factors, such as flagella, LPS, and exotoxin U in Pseudomonas aeruginosa [26].
Infection with human papillomavirus (HPV) is strongly correlated with malignant oral lesions. Yet, it may not be enough for the neoplastic transformation of normal epithelial cells. This could be explained by the high prevalence of HPV infections, the prolonged incubation period between initial HPV infection and the emergence of cancer, and the regression of most HPV-induced dysplasias. Chronic inflammation is suggested to promote oral carcinogenesis with HPV infection. Tumor necrosis factor α (TNFα) which is main mediator of chronic inflammation, reinforces inflammation-associated tumorigenesis by nuclear factor-κB signaling activation, which suppresses precancerous cells’ death through the evolution of inflammation-associated cancers in HPV infected cases [27]. The microbiome may offer signatures that can be used as potential biomarkers for early detection, discriminating oral cancer subtypes and predicting clinical behaviour as metastasis and oral cancer recurrences [28]. Using saliva may avoid biopsies and serves a non-invasive, inexpensive, and easily accessible diagnostic and prognosis tool in oral cancer patients [29].
ii Microbiome and Esophageal Cancer
a. Esophageal Adenocarcinoma
It is of high importance to better comprehend the function of the microbiome in esophageal adenocarcinoma. Previous studies used surgically resected samples and culture‐based techniques of esophageal adenocarcinoma and squamous cell carcinoma and found the same matching microbiota in both normal and cancerous tissues. Yet, these investigations were mainly interested in recognizing microbes related to infections postoperatively, not in confronting the non‐pathogenic bacteria in esophageal cancer patients and control cases, Narikiyo et al. lately used the 16S sequencing technique to distinguish microbiota of both normal and cancerous esophageal tissues [30-32]. Cancerous tissue specimens were collected from 20 patients who experienced surgical resection for cancer esophagus.
The oral periodontopathic spirochete Treponemadenticola, Streptococcus anginosus and Streptococcus mitis were dominant in both microbiota, but the pathological classification of the cancers was not given [33]. Blackett et al. used a culture-independent and mixed culture-dependent strategies to compare the microbiota in reflux-asymptomatic controls and patients with GERD, Barrett's esophagus, and esophageal adenocarcinoma. Significant enrichment of Campylobacter was noticed in Barrett's esophagus and GERD patient’s comparative with the controls and esophageal adenocarcinoma patients. Furthermore, cytokines involved in tumorigenesis (e.g. IL-18) were more up-regulated in tissues colonized with Campylobacter [34].
Considering the potential pathogenicity of Campylobacter species, its role in the emergence of esophageal adenocarcinoma may mimic the Helicobacter pylori role in cancer stomach [35]. The rapport between the esophageal adenocarcinoma development and microbiome has been investigated experimentally. Sawada et al. investigated whether altering microbiome with antibiotics would impact the emergence of esophageal adenocarcinoma in a rat model with esophagojejunostomy [36]. By using terminal restriction fragment length polymorphism analysis, the esophageal microbiomes were found to be different between the two groups; for example, the proportions of Lactobacillales were reduced in the antibiotics group, but Clostridium were elevated in the antibiotics group. Whilst the incidence of esophageal adenocarcinoma wasn’t impacted by the alteration of the microbiome. In a rat model with esophagojejunal anastomosis, Zaidi et al. reported a predominance of Escherichia coli in Barrett's esophagus and esophageal adenocarcinoma. This indicates an association between E. coli and TLR signaling pathway, furthermore, TLR 1-3, 6, 7 and 9 were notably up-regulated in esophageal adenocarcinoma comparative with normal epithelium, proposing that these early molecular changes are mediated by microbes in the rat model of esophageal adenocarcinoma tumorigenesis [37].
A significant correlation was found between transcriptionally active HPV, Barret’s esophagus, and esophageal adenocarcinoma, proposing a potential role in esophageal tumorigenesis. Moreover, when transcriptional activity markers (E6/E7 mRNA and p16INK4A) and HPV DNA were all positive, a very strong correlation between disease severity along the Barrett’s metaplasia-dysplasia-adenocarcinoma sequence was found [38].
b. Esophageal Squamous Cell Carcinoma
Esophageal microbiome, it is more prominent in adenocarcinoma when compares to squamous cell carcinoma (SCC). A study by Yu et al. reported an inverse relationship between the complexity of the microbiota population and the liability to develop squamous dysplasia [39]. Contamination of the esophageal epithelium with Porphyromonas gingivalis was not only implicated in the pathogenesis of esophageal SCC but also in the tumor cells differentiation and metastasis, suggesting it's potential role as a prognostic biomarker [40]. Gastric dysbiosis specially Erysipelotrichales and Clostridiales rich population is implicated in the progression of esophageal squamous cell carcinoma from dysplasia [41].
Altered salivary microbiome has been implicated in esophageal SCC, Beneficial microbiota as genera Lautropia, Bulleidia, Corynebacterium, Catonella, Peptococcus, Moryella and Cardiobacterium responsible for good oral health was less prominent in SCC when compared with healthy controls [42]. Fusobacterium nucleatum, oral microbiota that has been associated with multiple periodontal diseases. Previously linked to colon cancer, Fusobacterium nucleatum, has been abundantly found in 23% esophageal SCC in a study including 325 samples, recently linked to esophageal SCC [43].
Future detailed studies to determine different microbiome populations implicated in the development and progression of SCC paves the road for better understanding of esophageal SCC etiology and pathogenesis and developing de novo therapeutic and preventive strategies against esophageal SCC.
iii Microbiome and Gastric Cancer
a. H. pylori and Gastric Cancer
H. pylori have been adapted to live in the harsh, acidic environment of the stomach. These bacteria reduce its acidity so they can survive and change the environment around them. The spiral shape of H. pylori permits them to penetrate the stomach lining, where they are protected by mucus and thus the body’s immune cells are not able to reach them. The bacteria can inhibit the immune response resulting in several gastric problems [8]. The direct impact of H. pylori on tumorigenesis is exerted primarily through two virulence factors: CagA and VacA [44]. H. pylori is estimated to be responsible for 5.5% of all cancer cases and over 60% of gastric cancer cases [45].
Paik et al. found that H. pylori-induced expression of LC3, one of the major markers of autophagy, was more evident than that observed in the cells subjected to starvation for 48 h. Therefore, it is more possible that H. pylori may be one of the strong autophagy inducers like Nrf2 transcription factor that regulates the autophagic signaling pathway by modulating the adaptor protein p62/ SQSTM. These findings suggest the existence of a positive feedback loop between Nrf2 and p62/SQSTM1. One of the key target molecules of Nrf2 is the stress-responsive enzyme, HO-1. These findings elucidate the role of HO-1 in H. pylori-induced autophagy through Nrf2 activation [46].
β-catenin is a proto-oncogene that has been found to accumulate inside the nucleus in precancerous lesions of gastric cancer. H. pylori activates β-catenin expression, such that β-catenin activates its expression via H. pylori as a positive feedback mechanism to induce intestinal metaplasia [47].
b. Epstein-Barr Virus (EBV) and Gastric Cancer
Epstein-Barr virus (EBV) is involved in gastric carcinogenesis as about 9% of gastric tumors harbor monoclonal viral episomes. The Cancer Genome Atlas project identified EBV-positive tumors as one of four molecular subtypes of gastric cancer. EBV-positive tumors are characterized by recurrent PIK3CA mutation, absence of TP53 mutation, JAK2 amplification, and extreme DNA hypermethylation. These serologic patterns are consistent with viral reactivation in the presence of EBV positive tumors as three (anti-VCA, anti-EA-D, and anti-ZEBRA) of the four studied antibodies target proteins expressed during lytic replication. This pattern of viral reactivation could represent lytic phase infection in either EBV-positive gastric cancer cells or benign lymphocytes related to reduced immune function [48].
iv Microbiome and Hepatocellular Cancer
The role of the microbiome in hepatic tumorigenesis is mostly through inflammatory pathways that start by the interaction between the intestinal bacteria, liver, and immune system. The process mainly includes the interaction between Kupffer cells, macrophages, and PAMPs in the liver. In the cascade of eradicating microorganisms, most populations of Kupffer cells and macrophages respond to very low concentrations of PAMPs, endotoxins, or LPS via the activation of NF-κB by binding to TLRs, mostly TLR-4 and -9, and nucleotide-binding oligomerization domain-like receptor (NOD-like receptor). This accordingly leads to a chain of inflammatory reactions that induces inflammation and cytokine release [49, 50].
One study by Grat et al. investigated the intestinal microbiota (IM) of 15 patients with HCC undergoing liver transplantation and compared them to 15 patients who did not have HCC but had a similar etiology of cirrhosis and a similar stage. The study reported that HCC was strongly correlated with increased fecal Escherichia coli [51].
Another study assessed liver tissue samples in patients with HCC and found the presence of Helicobacter spp., proposing intestinal translocation as a potential mechanism for carcinogenesis. However, Helicobacter could not be found in patients with viral-induced HCC [52].
In another study, they introduced the dysbiosis (Ddys) index to measure dysbiosis and found that the Ddys was markedly boosted in patients with primary hepatocellular carcinoma comparative with the healthy controls. Additionally, the development of primary HCC is associated with the increase of the Ddys, although no significant difference was detected between patients with different stages of primary HCC, However, The increase in the bacterial genera that were closely correlated with dysbiosis in primary HCC occurs by Ddys. Further studies are needed to verify the application of this index for use in other diseases [53].
In a study by Pinero et al. Patients with HCC showed a more diverse gut microbiome than without-HCC group. First, HCC patients showed specific changes in family members of Firmicutes including a 5-fold decrease in family Leuconostocaceae and a 3-fold increase in abundance of Erysipelotrichaceae comparative with controls. Second, genus Fusobacterium was 5-fold less abundant in HCC patients comparative with controls. Third, the Bacteroides/Prevotella ratio showed an increase in patients with HCC due to the remarkable decrease in the genus Prevotella. This pattern has been correlated with an inflammatory medium and was recognized as HCC potential biomarkers, including genus Odoribacter and Butyricimonas of Odoribacteraceae family that showed an increased abundance, while Lachnospiraceae family genus Dorea showed a decreased abundance in HCC patients [54].
Hepato-tropic viruses, as HCV and HBV, are highly correlated with the emergence of HCC. Hepatic virus-induced hepatotumorigensis is usually a complex process that may comprise the induction of oxidative stress, cellular inflammation, and involvement of signaling pathways, resulting in selective activation of oncogenic pathways and integration of the viral genome into host DNA via host DNA deletion. Without entire viral elimination from the host, uncompromising replication stimulates inflammation that precipitates chronic liver disease and so resembles a risk factor for HCC. Limited information is available about the viruses involved in the dysbiosis of microbiome-mediated HCC. Yet, some studies have proposed that the pathophysiology of viral hepatitis is aggravated by the gut microbiota and may develop to advanced stages of HCC. The incorporation of the gut microbiome highly impacts the liver’s immune response, resulting in either viral eradication or persistence [55].
v Microbiome and Pancreatic Cancer
Patients with pancreatic ductal adenocarcinoma (PDAC) showed diverse microbiome changes comparative with healthy controls at several body sites, including oral cavity, pancreatic tissues, GIT using quantitative polymerase chain reaction (qPCR) and16S ribosomal RNA (16S rRNA) gene sequencing [56].
The human oral cavity is colonized by more than 700 different microorganism species, yet the oral microbiota remains relatively stable in healthy individuals [57]. Periodontitis is the most widely recognized infectious condition prompting tooth loss and has been correlated with different malignancies of the pancreas, colorectum and other extraintestinal organs [58-63]. Of the known independent risk factors for PDAC are Poor oral health status, pathogenic oral flora, periodontal disease, and tooth loss [64-70]. Microbiologists have reported the propagation of oral microbes to the pancreas by translocation or dissemination in both animal models and human subjects [71]. Many oral bacteria are involved in PDAC tumorigenesis such as Fusobacterium, Porphyromonas gingivalis (P. gingivalis), Streptococcus mitis (S. mitis) and Neisseria elongata (N. elongata) [72].
HCV and HBV are of the hepato-tropic viruses which cause hepatitis and hepatocellular carcinoma (HCC). Yet, their infection is not limited to the liver and these viruses can be found in extrahepatic tissues, including the pancreas. So, they may have a role in the tumorigenesis of extrahepatic cancers, including PDAC. HBsAg and HBcAg were detected in the cytoplasm of pancreatic acinar cells, and patients with chronic HBV infection showed partially elevated serum and urinary levels of pancreatic enzymes. Researchers detected HBsAg in the pancreatic juice of patients with HBV infection and was correlated with the emergence of chronic pancreatitis, proposing that HBV-related pancreatitis could be a precursor of PDAC. Agreed conclusions drawn from meta-analyses support the positive correlation of PDAC risk with HBV infection, especially for long-lasting persistent infection, chronic/inactive HBsAg carriers and occult infection. Apparently, HBV can replicate apart from infecting in the tumor and non-tumorous pancreatic tissues of PDAC patients. The REVEAL-HBV study reported the association between HBV and PDAC in subjects with higher viral DNA loads (HBV DNA > 300 copies/mL). In patients with HBV infection, researchers confirmed the integration of HBV DNA to pancreatic tissue and pancreatic metastases to the liver. HBV infection was reported to increase the rate of concurrent liver metastasis in patients with PDAC, which could be considered as a prognostic factor. However, some studies reported inconsistent conclusions regarding the correlation between PDAC and the presence of HBV and HCV [73].
vi Microbiome and Colorectal Cancer
Colorectal tumorigenesis is a complex process, affected by genetic and environmental factors with different causative mechanisms. Many of these mechanisms, such as inflammation, immune regulation, metabolism of dietary components and genotoxin production, are strongly related to the gut microbiota and have been comprehensively investigated. We will focus on selected microbiota-associated components in CRC tumorigenesis that are potential for modulation and clinical use [74]. The gut microbiota can influence colorectal tumorigenesis through many mechanisms, including microbial-related factors such as metabolites or genotoxins. Moreover, some bacteria can be directly procarcinogenic (driver bacteria) or proliferate as opportunistic microorganisms in the tumor-associated microenvironment (passenger bacteria) [75].
Some particular bacterial species can trigger incendiary reactions or produce poisons that straightforwardly harm gut cells. For example, Bacteroides fragilis and Enterococcus faecalis produce enterotoxins (for example fragylisin) and reactive oxygen species that cause oxidative DNA harm, prompt aggravation, and harm the epithelial hindrance [76]. Also, B. fragilis modulate the expression of β-catenin and affect cell multiplication [77]. Fusobacterium nucleatum can attack colonic epithelial cells by means of the FadA surface protein, which interfaces with E-cadherin to intervene changes in β-catenin and Wnt signaling [78].
Although the fecal microbiome is just an intermediary for the gut microbiome, this non-invasive methodology has provided vital data of the alterations in the gut biological system related to CRC and is the most regularly utilized inspecting technique in gut microbiome contemplates. Various investigations looking at fecal microbiota from CRC patients and few microbes, for example, Streptococcus gallolyticus, F. nucleatum, Escherichia coli, B. fragilis and E. faecalis, higher level in CRC patients when contrasted with the healthy control. Moreover, Roseburia, Clostridium, Faecalibacterium and Bifidobacterium are commonly lower in CRC patients [79].
By sequencing the V3 V4 region of the 16S rRNA gene, Sheng et al. analysed the fecal samples of healthy controls and patients with CRC. There was a marked variety of fecal gut microbiota in CRC patients compared with the healthy controls. There was a relative richness of Prevotella, Peptostreptococcus and Collinsella, but Escherichia and Shigella was markedly decreased comparative with healthy controls. Moreover, variance in the fecal gut microbiota was investigated in patients with stage I IV CRC and healthy controls. Stage I CRC patients showed marked increase in Collinsella, Peptostreptococcus and Ruminococcus compared to the healthy controls, while stage III CRC patients showed enrichment of Alistipes compared to patients with stage IV. Furthermore, another study found that the genera Coprobacter and Veillonella showed more abundance in the proximal segments than in the distal segments of the colon [80].
Among several bacterial candidates, F. nucleatum stood out as a key marker either when being investigated alone or combined with other bacteria specifically Clostridium symbiosum, C. hathewayi and colibactin-producing clbA+ bacteria. The faecal abundance of F. nucleatum can empower the use of faecal immunochemical test (FIT) to give higher sensitivity and specificity in detecting CRC than using FIT alone [75].
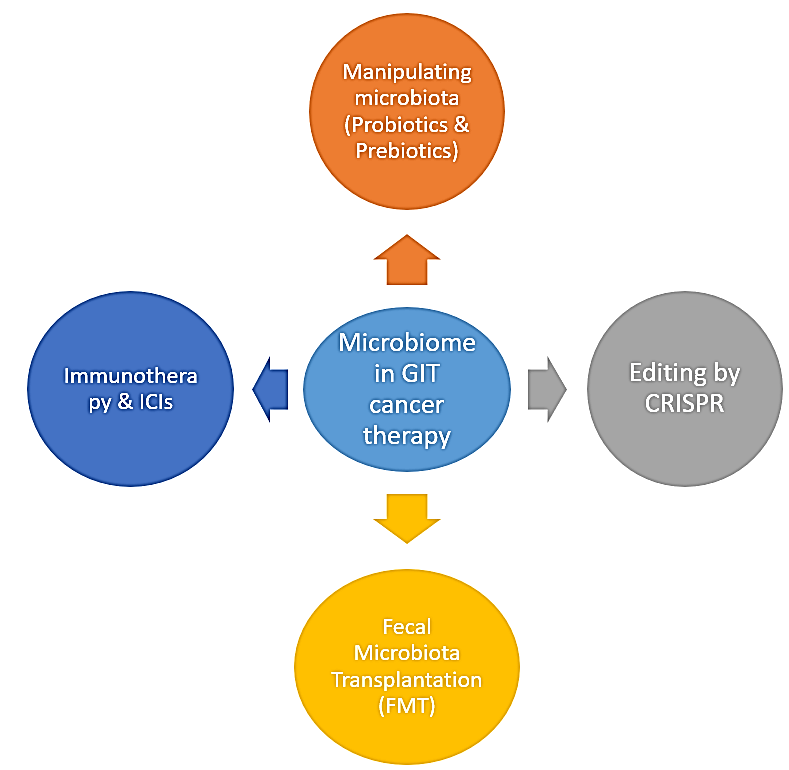
II Microbiome and Cancer Treatment (Figure 1)
Four unmistakable measures can be used to modify the impacts of the gut microbiota on anticancer treatment:
i. Anti-infection agents, synthetic concoctions with a particular cytotoxicity for at least one bacterial animal types.
ii. Probiotics, living microscopic organisms or different microorganisms that, when directed in satisfactory sums, give a wellbeing benefit.
iii. Prebiotics, no digestible exacerbates that invigorate the development and additionally elements of specific segments of the gut microbiota.
iv. Postbiotics, nonviable results or products of the gut microbiota that apply natural or biological activities in the host [81].
Figure 1: Role of microbiota and different modalities in GIT cancer therapy – Probiotics have been tried in animal tumor models for their capacity to prevent carcinogenesis with promising outcomes [87]. Song et al., have used nanoparticles loaded with spores of Bacillus coagulans (BC) for GIT cancer therapy [89]. Treatment with Lactobacillus rhamnosus significantly reduced the incidence of colorectal cancer in rats [90]. Prebiotics have been examined as potential method for inactivating colorectal malignancy [89]. Immunotherapy may change the gut microbiota which can affect interleukin dependent Th1 immune response which improve tumor control with sparing intestinal integrity [98]. Chmielewski et al. used T-cells redirected for universal cytokine-mediated killing (IL-18-inducing TRUCK construct) to develop a CEA-directed CAR T-cell that leads to enhanced antitumor activity in pancreatic cancer mice model [102]. The current experiments to engineer probiotic E. coli strains by CRISPR will be effectively new tools in cancer therapy [106]. Enabling the engineering of C. difficile by using the CRISPR tools could potentially be as an interesting probiotics [110]. Studies has been performed in mice to evaluate the efficacy of FMT in decreasing colon cancer with promising results [112].
III Manipulating the Microbiota for Cancer Therapy
Probiotics have been broadly tried in animal tumor models for their capacity to prevent carcinogenesis with promising outcomes. Also, genetically engineered probiotics have been effectively utilized as vectors for the delivery of tumor-associated antigens, immune-stimulatory molecules, or chemicals, enzymes, that utmost the toxic side effects of chemotherapy in animal models [82].
Song et al., have used nanoparticles loaded with spores of Bacillus coagulans (BC) for GIT cancer therapy. The spores can germinate to probiotics inside the intestine [83]. Saadat et al. used Kluyveromyces marxianus and Pichia kudriavzevii yeasts as probiotics to treat colon cancer through modulating AKT-1, mTOR, and JAK-1 pathways [84]. In a study by Huang et al., treatment with Lactobacillus rhamnosus significantly reduced the incidence of colorectal cancer in rats [85].
Although prebiotics (e.g. inulin or oligofructose) and postbiotics (e.g. butyrate) have been investigated as potential methods for inactivating colorectal malignancy, yet more studies are needed to reinforce these trials [84]. Furthermore, specific synthetic substances might be effectively used to contain the negative effect of the gut microbiota on the pharmacodynamics of specific chemotherapeutics. As a proof of rule, an intense inhibitor of bacterial (however not mammalian) β-glucuronidase has been appeared to shield mice from the intestinal side effects of irinotecan [86].
i Gut microbiota – Immunotherapy Crosstalk in GIT Cancer
The response to immunotherapy in GIT cancer depends on the following factors, genetic makeup of the tumor, how the immune system reacts to it, and regulatory mechanisms as the gut microbiota [81, 87-89]. Of note, two research groups highlight the role of gut microbiota in modulating the anticancer activity of CTLA4 and PD1 blockade. Vétizou et al. declared that the gut microbiota composition like the Bacteroides spp. can itself decrease the tumor volume and when combined with immunotherapy it more reduces the tumor size [90]. Furthermore, Lifestyle and immunotherapy may change the gut microbiota which can affect interleukin dependent Th1 immune response which improves tumor control both with sparing intestinal integrity [91]. Moreover, the oral administration of Bifidobacterium when combined with anti-PDL1 therapy nearly stops tumor growth [92]. Immune checkpoint inhibitors (ICIs) therapy is a novel procedure for cancer treatments. It is realized that heterogeneity of gut microbiome may bring about various results of treatment [93].
With the incorporation of genome-editing strategies such as CRISPR/Cas9 and transcription activator-like effector nucleases (TALENs), into synthetic biology, increasingly sophisticated and specific CAR T-cells (autologous T-cells re-directed towards a tumor-specific antigen) are being designed for clinical usage through knockout of a multiplex of inhibitory proteins like PD1, CTLA-4, and β-2 microglobulin, and intrinsic T-cell inhibitory enzymes in hematological and solid tumors [94]. Recently, Chmielewski et al. then used T-cells redirected for universal cytokine-mediated killing (IL-18-inducing TRUCK construct )to develop a CEA-directed CAR T-cell that leads to enhanced antitumor activity in pancreatic cancer mice model [95] .
ii Editing the Microbiome by CRISPR
While the results of the majority of therapeutic strategies that target the microbiota are usually not specific due to restricted comprehension of the ecological interactions which occur within microbes and with the host immune system. Clustered regularly interspaced short palindromic repeats (CRISPRs) – and CRISPR-associated (Cas) proteins present a powerful set of tools that will enhance the study of the microbiome and may lead to the development of new techniques to alter them [96].
It may now be recognized as a promising tool to modify probiotic strains for additional therapies [97]. Probiotic E. coli strains have been engineered to express specific antigens, antimicrobial compounds, sensing molecules that regulate pathogen virulence [98]. These recent experiments to modify probiotic E. coli strains will be effectively new tools in cancer therapy [99]. An innovative and potential strategy in the genome editing area is the use of the catalytic dead form of Cas9 (dCas9) fused to a cytosine deaminase or an adenosine deaminase at specific target positions, making the DNA break not required. Kondo et al. modified this technique for E. coli and were able to optimize up to 41 loci [100].
Bifidobacteria and Lactobacilli are the most frequently used probiotic bacteria aside from E. coli. Genetically-engineered Lactobacilli are being optimized as targeted therapies against many diseases including oral mucositis, inflammatory bowel disease and viral and bacterial infections [101]. Taking into consideration that Bifidobacterium is able to replicate in solid tumors, it offers the potential to genetically modify them to produce cancer inactivating compounds [102]. Interestingly, authorizing the engineering of C. difficile by using the CRISPR tools could potentially be an interesting probiotics [103]. Gene silencing with dCas9 (CRISPRi) is much easier to be conducted in bacteria than Cas9-mediated genome editing. This has resulted in the prompt and wide adoption of this strategy in various types of bacteria [104].
iii Fecal Microbiota Transplantation (FMT) in GIT Cancer
Some preclinical studies have been performed in mice to evaluate the efficacy of FMT in decreasing colon cancer with promising results, despite lack of control in this procedure because all gut microbiota is transferred with the therapeutic bacteria species [105].
Future Perspective
With increasing the precise characterization of single enteric, dysplastic, and immune cells by using high-throughput technologies like NGS sequencing and mass, many organisms have already been sequenced and become available in several databases. Thus, experimental validation of these preliminary results has become possible. Scientists shed light on “culturomics” to make more microbiota investigable, and translational bioinformatics implementations to facilitate efficient data analysis and integration. Recently, the development of three-dimensional cultures (organoids) has enabled the growth of normal intestinal units that has also been applied to study GIT diseases, gut microbe interactions, and colorectal cancer. Such organoids are amenable to gene editing by CRISPR and high-throughput small-molecule drug testing.
By throwing light on the molecular mechanisms involved in microbiome-based development and progression of GIT cancers, we hope to evolve unprecedented therapeutic strategies by modifying the microbiota. First, by modifying the combination of intestinal microbiota by prebiotics, probiotics, antibiotics, or microbiota transplants. Second, modulate the side effects and efficacy of available chemotherapy and immunotherapy using the manipulation of the microbiota. Furthermore, the microbiota may serve as a potential diagnostic or clinical outcome biomarker in GIT cancers if correct relationship between gut microbiota and poor clinical outcome has been established.
Conclusion
The host and the gut microbiota show a complex relationship that can be unbalanced in a state of gut microbiota dysbiosis secondary to environmental changes that lead to inflammation, modulation of immune checkpoint and GIT cancer. Comprehension of the microbiome composition and the different ways by which microbiome contributes to GIT tumorigenesis will offer new potentials for novel diagnostic, prognostic, and therapeutic tools.
Conflicts of Interest
None.
Article Info
Article Type
Review ArticlePublication history
Received: Mon 12, Oct 2020Accepted: Wed 25, Nov 2020
Published: Wed 16, Dec 2020
Copyright
© 2023 Maha Saad. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.JSO.2020.05.03
Author Info
Maha Saad Mansour Mohamed Kabash Aymn El Sayed Shafei Rawan Ellackany Ahmed Atef Mostafa Wael Marwa Matboli
Corresponding Author
Maha SaadBiochemistry Department, Faculty of Medicine, Modern University for Technology and Information, Cairo, Egypt
Figures & Tables
Table 1: Showing summary of microbiome involved in GIT cancers.
|
Disease |
Microbiome biomarker |
Reference |
|
Oral squamous cell carcinoma |
Fusobacterium periodonticum Prevotella melaninogenica Capnocytophaga gingivalis Streptococcus mitis Lactobacillaceae Lactobacillus Haemophilus Aggregatibacter Neisseria Gemellaceae Veillonella Porphyromonas Actinomyces Clostridium Enterobacteriaceae Pseudomonas Atopobium Campylobacter concisus Lautropia mirabilis Rothia dentocariosa Human papilloma virus (HPV) |
[17, 21-27] |
|
Esophageal adenocarcinoma |
Treponemadenticola Streptococcus anginosus Streptococcus mitis Campylobacter Clostridium Lactobacillales Escherichia coli HPV |
[33, 35, 37, 38] |
|
Esophageal squamous cell carcinoma |
Clostridiales Erysipelotrichales Porphyromonas gingivalis Lautropia, Bulleidia, Catonella, Corynebacterium, Moryella, Peptococcus Cardiobacterium Fusobacterium nucleatum |
[40-42, 43] |
|
Gastric cancer |
H. pylori Epstein-Barr virus (EBV) |
[8, 44, 45, 48] |
|
Hepatocellular cancer |
Helicobacter spp. Erysipelotrichaceae Leuconostocaceae Fusobacterium Prevotella Odoribacter Butyricimonas Dorea HBV HCV |
[52, 54, 55]
|
|
Pancreatic Cancer |
Porphyromonas gingivalis, Fusobacterium Neisseria elongata Streptococcus mitis HBV and HCV |
[72, 73] |
|
Colorectal Cancer |
Bacteroides fragilis Enterococcus faecalis Fusobacterium nucleatum Streptococcus gallolyticus Escherichia coli Roseburia Clostridium Faecalibacterium Bifidobacterium Collinsella Peptostreptococcus Prevotella Shigella Ruminococcus Alistipes Veillonella Coprobacter Clostridium symbiosum C. hathewayi |
[75, 76, 78-80] |
References
- Collins D, Hogan AM, Winter DC (2011) Microbial and viral pathogens in colorectal cancer. Lancet Oncol 12: 504-512. [Crossref]
- Stojanovic MR, Vos WMD (2014) The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38: 996-1047. [Crossref]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O et al. (2014) Human genetics shape the gut microbiome. Cell 159: 789-99. [Crossref]
- Proal AD, Albert PJ, Marshall TG (2013) The human microbiome and autoimmunity. Curr Opin Rheumatol 25: 234-240. [Crossref]
- Belizário JE, Napolitano M (2015) Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front Microbiol 6: 1050. [Crossref]
- Cresci GA, Izzo K (2019) Gut Microbiome. InAdult Short Bowel Syndrome. Academic Press 45-54.
- Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ (2015) Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis 26: 26191. [Crossref]
- Colledge H, Cafasso J (2017) H. pylori Infection. Healthline.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA et al. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68: 394-424. [Crossref]
- Klampfer L (2011) Cytokines, inflammation and colon cancer. Curr Cancer Drug Targets 11: 451-464. [Crossref]
- Hattori N, Ushijima T (2016) Epigenetic impact of infection on carcinogenesis: mechanisms and applications. Genome Med 8: 10. [Crossref]
- Putoczki TL, Thiem S, Loving A, Busuttil RA, Wilson NJ et al. (2013) Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell 24: 257-271. [Crossref]
- Hussain SP, Amstad P, Raja K, Ambs S, Nagashima M et al. (2000) Increased p53 mutation load in noncancerous colon tissue from ulcerative colitis: a cancer-prone chronic inflammatory disease. Cancer Res 60: 3333-3337. [Crossref]
- Gensollen T, Iyer SS, Kasper DL, Blumberg RS (2016) How colonization by microbiota in early life shapes the immune system. Science 352: 539-544. [Crossref]
- Brown DG, Rao S, Weir TL, O'Malia J, Bazan M et al. (2016) Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab 4: 11. [Crossref]
- Schmidt BL, Kuczynski J, Bhattacharya A, Huey B, Corby PM et al. (2014) Changes in abundance of oral microbiota associated with oral cancer. PLoS One 9: e98741. [Crossref]
- Vergara D, Simeone P, Damato M, Maffia M, Lanuti P et al. (2019) The Cancer Microbiota: EMT and Inflammation as Shared Molecular Mechanisms Associated with Plasticity and Progression. J Oncol 2019: 1253727. [Crossref]
- Multhoff G, Molls M, Radons J (2012) Chronic inflammation in cancer development. Front Immunol 2: 98. [Crossref]
- Sztukowska MN, Ojo A, Ahmed S, Carenbauer Al, Wang Q et al. (2016) Porphyromonas gingivalisinitiates a mesenchymal-like transition through ZEB1 in gingival epithelial cells. Cell Microbiol 18: 844-858. [Crossref]
- Ha NH, Woo BH, Kim DJ, Ha ES, Choi J II et al. (2015) Prolonged and repetitive exposure to Porphyromonas gingivalis increases the aggressiveness of oral cancer cells by promoting the acquisition of cancer stem cell properties. Tumor Biol 36: 9947-9960. [Crossref]
- Mager DL, Haffajee AD, Devlin PM, Norris CM, Posner MR et al. (2005) The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, nonrandomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med 3: 27. [Crossref]
- Mager DL (2006) Bacteria and cancer: cause, coincidence or cure? A review. J Transl Med 4: 14. [Crossref]
- Hooper SJ, Crean SJ, Lewis MAO, Spratt DA, Wade WG et al. (2006) Viable bacteria present within oral squamous cell carcinoma tissue. J Clin Microbiol 44: 1719-1725. [Crossref]
- Hooper SJ, Crean SJ, Fardy MJ, Lewis MAO, Spratt DA et al. (2007) A molecular analysis of the bacteria present within oral squamous cell carcinoma. J Med Microbiol 56: 1651-1659. [Crossref]
- Nagy KN, Sonkodi I, Szöke I, Nagy E, Newman HN (1998) The microflora associated with human oral carcinomas. Oral Oncol 34: 304-308. [Crossref]
- Perera M, Hebshi NNA, Perera I, Ipe D, Ulett GC et al. (2018) Inflammatory bacteriome and oral squamous cell carcinoma. J Dent Res 97: 725-732. [Crossref]
- Hong HS, Akhavan J, Lee SH, Kim RH, Kang MK et al. (2020) Proinflammatory cytokine TNFα promotes HPV-associated oral carcinogenesis by increasing cancer stemness. Int J Oral Sci 12: 3. [Crossref]
- Markopoulos AK, Michailidou EZ, Tzimagiorgis G (2010) Salivary markers for oral cancer detection. Open Dent J 4: 172-178. [Crossref]
- He J, Li Y, Cao Y, Xue J, Zhou X (2015) The oral microbiome diversity and its relation to human diseases. Folia Microbiol 60: 69-80. [Crossref]
- Lau WF, Wong J, Lam KH, Ong GB (1981) Oesophageal microbial flora in carcinoma of the oesophagus. Aust N Z J Surg 51: 52-55. [Crossref]
- Finlay IG, Wright PA, Menzies T, McArdle CS (1982) Microbial flora in carcinoma of oesophagus. Thorax 37: 181-184. [Crossref]
- Mannell A, Plant M, Frolich J (1983) The microflora of the oesophagus. Ann R Coll Surg Engl 65: 152-154. [Crossref]
- Narikiyo M, Tanabe C, Yamada Y, Igaki H, Tachimori Y et al. (2004) Frequent and preferential infection of Treponemadenticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Sci 95: 569-574. [Crossref]
- Blackett KL, Siddhi SS, Cleary S, Steed H, Milleret MH et al. (2013) Oesophageal bacterial biofilm changes in gastro‐oesophageal reflux disease, Barrett's and oesophageal carcinoma: association or causality? Aliment Pharmacol Ther 37: 1084-1092. [Crossref]
- Man SM (2011) The clinical importance of emerging Campylobacter species. Nat Rev Gastroenterol Hepatol 8: 669-685. [Crossref]
- Sawada A, Fujiwara Y, Nagami Y, Tanaka F, Yamagami H et al. (2016) Alteration of esophageal microbiome by antibiotic treatment does not affect incidence of rat esophageal adenocarcinoma. Dig Dis Sci 61: 3161-3168. [Crossref]
- Zaidi AH, Kelly LA, Kreft RE, Barlek M, Omstead AN et al. (2016) Associations of microbiota and toll‐like receptor signaling pathway in esophageal adenocarcinoma. BMC Cancer 16: 52. [Crossref]
- Haddad SA, Zimaity HE, Bakhtiari SH, Rajendra S, Streutker CJ et al. (2014) Infection and esophageal cancer. Ann N Y Acad Sci 1325: 187-196. [Crossref]
- Yu G, Gail MH, Shi J, Ceraj VK, Paster BJ et al. (2014) Association between upper digestive tract microbiota and cancer‐predisposing states in the esophagus and stomach. Cancer Epidemiol Biomarkers Prev 23: 735-741. [Crossref]
- Gao S, Li S, Ma Z, Shuo Liang, Tanyou Shan et al. (2016) Presence of Porphyromonasgingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect Agent Cancer 11: 3. [Crossref]
- Nasrollahzadeh D, Malekzadeh R, Ploner A, Shakeri R, Sotoudeh M et al. (2015) Variations of gastric corpus microbiota are associated with early esophageal squamous cell carcinoma and squamous dysplasia. Sci Rep 5: 8820. [Crossref]
- Chen X, Yuan Z, Lu M, Zhang Y, Jin L et al. (2017) Poor oral health is associated with an increased risk of esophageal squamous cell carcinoma ‐ a population‐based case‐control study in China. Int J Cancer 140: 626-635. [Crossref]
- Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K et al. (2016) Human microbiome fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res 22: 5574-5581. [Crossref]
- Yousefi B, Eslami M, Kokhaei P, Valizadeh S, Ghasemian A (2019) Role of autophagy associated with Helicobacter pylori CagA and VacA toxins in gastric cancer. Koomesh 21: 2015-214.
- Loor A, Dumitraşcu DL (2016) Helicobacter pylori infection, gastric cancer and gastropanel. Rom J Intern Med 54: 151-156. [Crossref]
- Paik JY, Lee HG, Piao JY, Kim SJ, Kim DH et al. (2019) Helicobacter pylori infection promotes autophagy through Nrf2-mediated heme oxygenase upregulation in human gastric cancer cells. Biochem Pharmacol 162: 89-97. [Crossref]
- Alfarouk KO, Bashir AHH, Aljarbou AN, Ramadan ARM, Muddathir AK et al. (2019) The Possible Role of Helicobacter pylori in Gastric Cancer and Its Management. Front Oncol 9: 75. [Crossref]
- Camargo MC, Kim KM, Matsuo K, Torres J, Liao LM et al. (2020) Circulating Antibodies against Epstein–Barr Virus (EBV) and p53 in EBV-Positive and-Negative Gastric Cancer. Cancer Epidemiol Biomarkers Prev 29: 414-419. [Crossref]
- Chassaing B, Mesmin LE, Gewirtz AT (2014) Microbiota‐liver axis in hepatic disease. Hepatology 59: 328-339. [Crossref]
- X Tao, N Wang, W Qin (2015) Gut Microbiota and Hepatocellular Carcinoma. Gastrointest Tumors 33-40. [Crossref]
- Grat M, Wronka KM, Krasnodebski M, Masior L, Lewandowski Z et al. (2016) Profile of Gut Microbiota Associated With the Presence of Hepatocellular Cancer in Patients With Liver Cirrhosis. Transplant Proc 48: 1687-1691. [Crossref]
- Kruttgen A, Horz HP, Heynemann JW, Vucur M, Trautwein C et al. (2012) Study on the association of Helicobacter species with viral hepatitis-induced hepatocellular carcinoma. Gut Microbes. [Crossref]
- Ni J, Huang R, Zhou H, Xu X, Li Y et al. (2019) Analysis of the relationship between the degree of dysbiosis in gut microbiota and prognosis at different stages of primary hepatocellular carcinoma. Front Microbiol 10: 1458. [Crossref]
- Piñero F, Vazquez M, Baré P, Rohr C, Mendizabal M et al. (2019) A different gut microbiome linked to inflammation found in cirrhotic patients with and without hepatocellular carcinoma. Ann Hepatol 18: 480-487. [Crossref]
- Gupta H, Youn GS, Shin MJ, Suk KT (2019) Role of gut microbiota in hepatocarcinogenesis. Microorganisms 7: 121. [Crossref]
- Archambault EN, Keim P, Hoff VD (2017) Microbiome and pancreatic cancer: A comprehensive topic review of literature. World J Gastroenterol 23: 1899-1908.
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR et al. (2010) The human oral microbiome. J Bacteriol 192: 5002-5017. [Crossref]
- Michaud DS, Izard J, Benartzi CSW, You DH, Grote VA et al. (2013) Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut 62: 1764-1770. [Crossref]
- Tezal M, Sullivan MA, Hyland A, Marshall JR, Stoler D et al. (2009) Chronic periodontitis and the incidence of head and neck squamous cell carcinoma. Cancer Epidemiol Biomark Prev 18: 2406-2412. [Crossref]
- Mai X, LaMonte MJ, Hovey KM, Nwizu N, Freudenheim JL et al. (2014) History of periodontal disease diagnosis and lung cancer incidence in the Women's Health Initiative Observational Study. Cancer Causes Control 25: 1045-1053. [Crossref]
- Urbaniak C, Cummins J, Brackstone M, Macklaim JM, Gloor GB et al. (2014) Microbiota of human breast tissue. Appl Environ Microbiol 80: 3007-3014. [Crossref]
- Freudenheim JL, Genco RJ, LaMonte MJ, Millen AE, Hovey KM et al. (2016) Periodontal Disease and Breast Cancer: Prospective Cohort Study of Postmenopausal Women. Cancer Epidemiol Biomark Prev 25: 43-50. [Crossref]
- Nwizu NN, Marshall JR, Moysich K, Genco RJ, Hovey KM et al. (2017) Periodontal Disease and Incident Cancer Risk among Postmenopausal Women: Results from the Women's Health Initiative Observational Cohort. Cancer Epidemiol Biomark Prev 26: 1255-1265. [Crossref]
- Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D et al. (2012) Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61: 582-588. [Crossref]
- Hujoel PP, Drangsholt M, Spiekerman C, Weiss NS (2003) An exploration of the periodontitis-cancer association. Ann Epidemiol 13: 312-316. [Crossref]
- Michaud DS, Joshipura K, Giovannucci E, Fuchs CS (2007) A prospective study of periodontal disease and pancreatic cancer in US male health professionals. J Natl Cancer Inst 99: 171-175. [Crossref]
- Ahn J, Segers S, Hayes RB (2012) Periodontal disease, Porphyromonasgingivalis serum antibody levels and orodigestive cancer mortality. Carcinogenesis 33: 1055-1058. [Crossref]
- Meyer MS, Joshipura K, Giovannucci E, Michaud DS (2008) A review of the relationship between tooth loss, periodontal disease, and cancer. Cancer Causes Control 19: 895-907. [Crossref]
- Stolzenberg Solomon RZ, Dodd KW, Blaser MJ, Virtamo J, Taylor PR et al. (2003) Tooth loss, pancreatic cancer, and Helicobacter pylori. Am J Clin Nutr 78: 176-181. [Crossref]
- Hiraki A, Matsuo K, Suzuki T, Kawase T, Tajima K (2008) Teeth loss and risk of cancer at 14 common sites in Japanese. Cancer Epidemiol Biomark Prev 17: 1222-1227. [Crossref]
- Michaud DS, Liu Y, Meyer M, Giovannucci E, Joshipura K (2008) Periodontal disease, tooth loss, and cancer risk in male health professionals: a prospective cohort study. Lancet Oncol 9: 550-558. [Crossref]
- Hayashi C, Gudino CV, Gibson FC 3rd, Genco CA (2010) Review: Pathogen-induced inflammation at sites distant from oral infection: bacterial persistence and induction of cell-specific innate immune inflammatory pathways. Mol Oral Microbiol 25: 305-316. [Crossref]
- Wei MY, Shi S, Liang C, Meng QC, Hua J et al. (2019) The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol Cancer 18: 97. [Crossref]
- Tilg H, Adolph TE, Gerner RR, Moschen AR (2018) The intestinal microbiota in colorectal cancer. Cancer Cell 33: 954-964. [Crossref]
- Wong SH, Yu J (2019) Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol16: 690-704. [Crossref]
- Cliffe MJ, Goodwin AL (2012) PASCal: a principal axis strain calculator for thermal expansion and compressibility determination. J Appl Crystallogr 45: 1321-1329.
- Wu S, Morin PJ, Maouyo D, Sears CL (2003) Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 124: 392-400. [Crossref]
- Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A et al. (2018) Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell 33: 125.e3 -136.e3. [Crossref]
- Saus E, Iraola Guzmán S, Willis JR, Brunet Vega A, Gabaldón T (2019) Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Mol Aspects Med 69: 93-106. [Crossref]
- Sheng Q, Du H, Cheng X, Cheng X, Tang Y et al. (2019) Characteristics of fecal gut microbiota in patients with colorectal cancer at different stages and different sites. Oncol Lett 18: 4834-4844. [Crossref]
- Zitvogel L, Galluzzi L, Viaud S, Vétizou M, Daillère R et al. (2015) Cancer and the gut microbiota: an unexpected link. Sci Transl Med 7: 271ps1. [Crossref]
- Zhao H, Zhang F, Chai J, Wang J (2020) Effect of lactic acid bacteria on Listeria monocytogenes infection and innate immunity in rabbits. Czech J Anim Sci 65: 23-30.
- Song Q, Zheng C, Jia J, Zhao H, Feng Q et al. (2019) A Probiotic Spore‐Based Oral Autonomous Nanoparticles Generator for Cancer Therapy. Adv Mater 31: e1903793. [Crossref]
- Saadat YR, Khosroushahi AY, Movassaghpour AA, Talebi M, Gargari BP (2020) Modulatory role of exopolysaccharides of Kluyveromyces marxianus and Pichia kudriavzevii as probiotic yeasts from dairy products in human colon cancer cells. J Functional Foods 64: 103675.
- Huang J, Wang D, Zhang A, Zhong Q, Huang Q (2019) Lactobacillus rhamnosus confers protection against colorectal cancer in rats. Tropic J Pharmaceut Res 18.
- Cheng KW, Tseng CH, Tzeng CC, Leu YL, Cheng TC et al. (2019) Pharmacological inhibition of bacterial β-glucuronidase prevents irinotecan-induced diarrhea without impairing its antitumor efficacy in vivo. Pharmacol Res 139: 41-49. [Crossref]
- Hold GL (2016) Gastrointestinal microbiota and colon cancer. Dig Dis 34: 244-250. [Crossref]
- Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R et al. (2013) The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342: 971-976. [Crossref]
- Hefazi M, Patnaik MM, Hogan WJ, Litzow MR, Pardi DS et al. (2017) Safety and efficacy of fecal microbiota transplant for recurrent clostridium difficile infection in patients with cancer treated with cytotoxic chemotherapy: a single-institution retrospective case series. Mayo Clin Proc 92: 1617-1624. [Crossref]
- Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N et al. (2015) Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350: 1079-1084. [Crossref]
- Procaccio L, Schirripa M, Fassan M, Vecchione L, Bergamo F et al. (2017) Immunotherapy in Gastrointestinal Cancers. Cancer Immunother Identific Progn Predict Biomarkers 2017: 17.
- Lote H, Cafferkey C, Chau I (2015) PD-1 and PD-L1 blockade in gastrointestinal malignancies. Cancer Treat Rev 41: 893-903. [Crossref]
- Chilakapati SR, Ricciuti J, Zsiros E (2020) Microbiome and cancer immunotherapy. Curr Opin Biotechnol 65: 114-117.
- Abid MB, Shah NN, Maatman TC, Hari PN (2019) Gut microbiome and CAR-T therapy. Exp Hematol Oncol 8: 31. [Crossref]
- Chmielewski M, Abken H (2017) CAR T cells releasing IL-18 convert to T-Bet(high) FoxO1(low) effectors that exhibit augmented activity against advanced solid tumors. Cell Rep 21: 3205-3219. [Crossref]
- Ramachandran G, Bikard D (2019) Editing the microbiome the CRISPR way. Philos Trans R Soc Lond B Biol Sci 374: 20180103. [Crossref]
- Bober JR, Beisel CL, Nair NU (2018) Synthetic biology approaches to engineer probiotics and members of the human microbiota for biomedical applications. Annu Rev Biomed Eng 20: 277-300. [Crossref]
- Duan F, March JC (2010) Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc Natl Acad Sci U S A 107: 11260-11264. [Crossref]
- Singh B, Mal G, Marotta F (2017) Designer probiotics: paving the way to living therapeutics. Trends Biotechnol 35: 679-682. [Crossref]
- Banno S, Nishida K, Arazoe T, Mitsunobu H, Kondo A (2018) Deaminase-mediated multiplex genome editing in Escherichia coli. Nat Microbiol 3: 423-429. [Crossref]
- Oh JH, van Pijkeren JP (2014) CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res 42: e131. [Crossref]
- Fujimori M (2006) Genetically engineered bifidobacterium as a drug delivery system for systemic therapy of metastatic breast cancer patients. Breast Cancer 13: 27-31. [Crossref]
- Woo TDH, Oka K, Takahashi M, Hojo F, Osaki T et al. (2011) Inhibition of the cytotoxic effect of Clostridium difficile in vitro by Clostridium butyricum MIYAIRI 588 strain. J Med Microbiol 60: 1617-1625. [Crossref]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS et al. (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173-1183. [Crossref]
- Bel S, Elkis Y, Elifantz H, Koren O, Ben Hamo R et al. (2014) Reprogrammed and transmissible intestinal microbiota confer diminished susceptibility to induced colitis in TMF−/− mice. Proc Natl Acad Sci U S A 111: 4964-4969. [Crossref]
- Almeqdadi M, Mana MD, Roper J, Yilmaz ÖH (2019) Gut organoids: mini-tissues in culture to study intestinal physiology and disease. Am J Physiol Cell Physiol 317: C405-C419. [Crossref]
- Meng C, Bai C, Brown TD, Hood LE, Tian Q (2018) Human Gut Microbiota and Gastrointestinal Cancer. Genomics Proteomics Bioinformatics 16: 33-49. [Crossref]
