Serum and Stool miR-135b Levels as a Potential Diagnostic Biomarker for Colorectal Cancer
A B S T R A C T
Background: By considering the high incidence and mortality rate of colorectal cancer (CRC), finding the noninvasive biomarker for detection of patients with cancer is the main purpose of more and more cancer studies.
Methods: In this research, the expression level of miR-135b in serum and stool of patients with colorectal cancer was investigated as a diagnostic marker. Using the real-time PCR, the relative expression level of miR-135b in serum and stool in 40 patients with colorectal cancer, paired with 40 healthy controls, was determined. Then its sensitivity and specificity were rated, via ROC curves analysis.
Results: Expression levels of miR-135b in serum and stool of CRC patients were 32.4 and 15.7 times higher in serum and stool, compared to that of healthy control respectively (P<0.05). ROC curves analysis exhibited that serum miR-135b levels were powerful in detecting CRC patients from control subjects, with a sensitivity of 92.7% and a specificity of 89.7% (AUC: 0.929). In addition, stool miR-135b levels strongly distinguished CRC patients from control subjects with a sensitivity of 92.7% and a specificity of 87.2% (AUC: 0.919).
Conclusions: The results of the current study indicate that serum and stool miR-135b expression levels seem to be used as a potential diagnostic biomarker for CRC patients. However further studies with large sample size are needed for approving the miR-135b as a noninvasive diagnostic biomarker of CRC.
Keywords
Serum, stool, miR-135b, biomarker, colorectal neoplasm
Introduction
Colorectal cancer is one of the most commonly diagnosed malignancies in human and is the second leading cause of cancer death [1]. TNM staging is considered as a standard method in routine use to determine CRC prognosis, in patients with high risk of mortality who could also respond to treatment [2]. Recently, molecular techniques, especially gene expression profiling, have been used increasingly in order to improve CRC classification [3, 4]. CRC is a multistep process characterized by genetic and epigenetic alterations that affect the main cellular pathways involved in growth and development [5]. Moreover, a better understanding of the molecular mechanisms involved in CRC development may contribute to exploit potential helpful prognostic biomarker and therapeutic target for CRC [6]. Early detection of cancer improves survival and therefore, the search for early detection biomarkers is highly warranted [7, 8]. In recent years, looking for prognostic biomarkers in CRC has increased and as a result, different miRNA expression analysis in tumor tissue studies also soared [9-11].
There are several interesting candidates, but the different finding makes it difficult to draw undoubted conclusions [12]. In addition to being found in tissue, miRNAs have also stably been detected in feces and serum making them potential good biomarkers [13-16].
MicroRNAs (miRNAs) are a class of short, non-coding single stranded RNA molecules (20-24 nucleotides) that regulate gene expression by post-transcriptional silencing of target mRNAs (mRNAs) through complementary binding and they also lead to translation of some target mRNAs [17-20]. In humans, miRNAs play a substantial role in the regulation of biologic processes such as development, differentiation, cell proliferation and apoptosis [17, 18, 21]. Recent studies indicated that dysregulation of miRNA is the hallmark of tumorigenesis so, miRNAs can be used as a diagnostic biomarker of cancer [19, 21, 22].
MiR-135b is overexpressed in adenomas and adenocarcinomas and one of its targets is APC, implying that the upregulation of miR-135b is an early event in CRC and might be used as an early detection biomarker [23, 24]. Adenomatous Polyposis Coli (APC) is a tumor suppressor gene that regulates beta-catenin concentrations and its protein interacts with E-cadherin [1, 10]. Mutations in the APC gene, leading to reduced expression, are defined as an early event in CRC [25, 26]. The aim of this research was to evaluate miR-135b expression through a quantitative reverse transcription PCR (qPCR) in CRC, to investigate its clinical significance and to evaluate the potential usefulness of miR-135b as a diagnostic biomarker.
Table 1: Patient’s clinical characteristics.
|
Healthy Controls |
Patients |
|
|
|
18 22 |
16 24 |
≤50 |
Age |
|
>50 |
|||
|
21 |
21 |
Male |
Genus |
|
19 |
19 |
Female |
|
|
|
11 |
I |
TNM staging |
|
|
16 |
II |
|
|
|
6 |
III |
|
|
|
7 |
IV |
|
|
|
32 |
Right side Colon |
Tumor location |
|
|
18 |
Left side colon |
These Patient Demographics information were extracted from patients Files in Shariati hospital.
Methods
I Participants and Sample Collection
This study was based on 40 patients and 40 control samples collected from Shariati hospital, Tehran, Iran, from 2014 to 2015 (Table 1). All processes were carried out with advised consent and approved by the ethical committee for clinical research at the hospital. Exclusion and inclusion criteria for the patients and controls were regarded throughout the study (Table 2).
Table 2: Inclusion and Exclusion Criteria to undergoing the study.
|
|
Patient |
Healthy control |
|
Inclusion Criteria |
A person that Was confirmed as malignant or precancerous by colonoscopy and histopathologic examination and was agreed to undergo the study. |
|
|
Exclusion Criteria |
A person who:
|
A person who:
|
Patients and healthy control group based on these criteria Were enrolled and the most of case histories were extracted from patient records.
II Serum and Stool Collection and RNA Extraction
Whole blood and stool samples of Patients and control group were collected after colonoscopy, and before surgery. Whole blood samples were collected in 5ml tubes (RNase-free) and were left to clot at room temperature for 30 minutes then they were centrifuged at 2000 rpm at 4°C for 10 minutes, The serum was then separated, aliquoted and stored at -80°C until needed. Stool samples were collected freshly and immediately flash frozen in liquid nitrogen and stored at -80 ◦C until used. Demographic and clinical information documented at the time of sampling included the stage, type, and grade of the tumor. Tumors were staged in accordance with American Joint Committee on Cancer (AJCC) TNM staging [13, 14]. One of the very important impacts on the results is the quality of total RNA, which was extracted from prototype [27]. Therefore, in this study, miRNeasy serum/plasma kit (Cat number: Q217184; Qiagen, Germany) for serum samples and miRNeasy mini kit (Cat number: Q217004; Qiagen, Germany) for stool were used, that are most qualified methods for miRNAs extraction [28]. Total miRNAs were extracted in accordance with the manufacturer’s instructions, and the concentration and purity of separated miRNA were assayed by Bio photometer system (Eppendorf AG, Hamburg, Germany).
III cDNA Synthesis and Real-Time PCR
For the cDNA synthesis and real-time PCR, the LNA kits and primers of Exiqon company were used that are totally exclusive to miRNA amplification [29]. CDNA synthesis was done by miRCURY LNA Universal cDNA synthesis kit II (Cat number: 203301; Exiqon, Vedbaek, Denmark). Real-time PCR was done by Exilent Sybergreen master mix (Cat number: 203403; Exiqon, Germany) and miR-16, RNU6B and miR-135b specific LNA™ PCR primer sets (Exiqon) according to manufacturer’s instructions. The real-time PCR condition was initial denaturation at 95 °C for 10 min, followed by 48 cycles of denaturation at 95 °C for 10 s (40 cycles for manufacturer), annealing and extension at 60 °C for 1 min and ramp-rate 1.6°C/s, in CFX96 real-time PCR system (Bio-Rad, Milan, Italy). The specificity of the PCR products were verified by melting curve analysis after last amplification procedure. To assure reproducibility and uniformity of the results, for all of the samples, real-time PCR reaction was done duplicated. In a pilot study, before normalization step, U6 small nuclear RNA (RNU6B) and miR-16 expression were examined as a reference gene, because a faultless internal control for stool-based research of miRNA, has not yet been approved. Due to the unchanging expression level, miR-16 was preferred as reference housekeeping gene for data normalization in serum and stool examinations.
IV Statistical Analysis
The efficiency of real-time PCR for both miR-135b and miR-16 was about 2; so, the rate of upregulation was measured using ΔΔCT methods, by REST (Relative Expression Software Tool) 2009 (Qiagen, Hilden, Germany) based on the relative expression of a miR-135b versus miR-16 as a reference gene [30]. The computing of -ΔΔCT was accomplished via transform menu of IBM SPSS statistics® version 20 (Chicago, IL, USA). For the analyses of variance, Kruskal-Wallis and Mann-Whitney statistics test were used to evaluate differences in serum or stool miRNA expression. Receiver operating characteristic (ROC) analysis was performed to determine the sensitivity and specificity of miR-135b expression in serum and stool of CRC patients as a diagnostic test. Data analysis was done with MedCalc®version 13.1.2.0-64bit (Acacialaan 22, 8400 Ostend, Belgium).
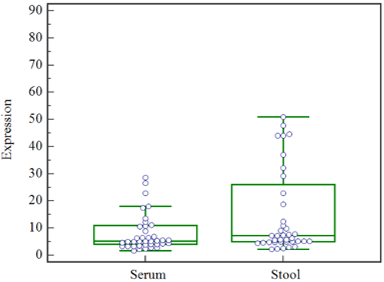
Figure 1: The expression levels of miR-135b in serum and stool. The results of Mann-Whitney test show that, In the CRC patients, the expression levels of miR-135b in serum compared to stool was not different significantly(P<0.05).
Results
As a primary step, we determined whether miR-135b in stool and serum of colorectal cancer patient in comparison with normal control is upregulated. MiR-135b in serum and stool were upregulated in CRC patients by 32.4 times in serum and 15.7 times, in the stool, compared to the normal serum and stool (P<0.05). The results of Mann-Whitney test show that the expression levels of miR-135b in serum and stool was significantly associated (P<0.05) (Figure 1).
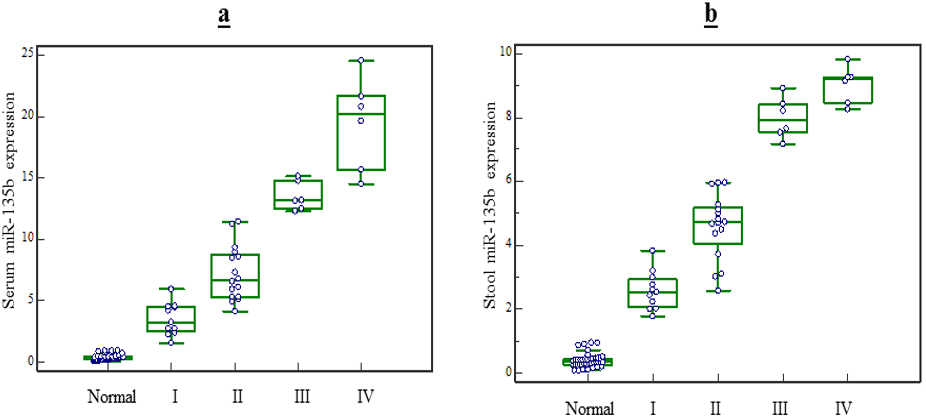
Figure 2: MiR-135b expression level in serum and stool based on patients clinical TNM stages. The result of Kruskal-Wallis test showed: miR-135b expression level in serum and stool based on patients clinical TNM stages. a) The expression level of miR-135b in serum can be hold as diagnostic marker to detect the TNM stage of CRC patients(P<0.05). b) The expression level of miR-135b in stool can be hold as marker to detect the TNM stage III, IV from I or II(P<0.05) but is unable to distinguish stage III from IV (P>0.05).
I MiR-135b Expression Level in Serum and Stool, Based on Patients Clinical Characteristics
Based on TNM staging, the expression level of miR-135b in serum raised respectively with stages, and expression levels in stage IV patients significantly elevated compared to stage I, II or III patients (p<0.05 for all stages). Nevertheless, stool expression levels in stage IV patients did not significantly elevate compared to stage III patients (p>0.05), but it raised compared to stages I or II (p<0.05) (Figure 2). on the other hand, the expression level of miR-135b in serum and stool of CRC patients did not vary with age, genus, tumor type, tumor size and tumor location (p>0.05) (Table 3).
Table 3: MiR-21 expression level in serum and stool, based on patients’ clinical characteristics.
|
|
|
MiR-21 in serum |
MiR-21 in stool |
||
|
Average rank |
P value |
Average rank |
P value |
||
|
Age |
≤50 |
20.37 |
0.688(>0.05) |
17.25 |
0.151(>0.05) |
|
>50 |
19.90 |
22.67 |
|||
|
Genus |
Male |
19.81 |
0.694(>0.05) |
20.52 |
0.989(>0.05) |
|
Female |
21.26 |
20.47 |
|||
|
Tumor type |
Colon |
21.09 |
0.52(>0.05) |
20.33 |
0.852(>0.05) |
|
Rectum |
18.12 |
20.19 |
|||
|
Tumor size |
≤5cm |
18.88 |
0.38 (>0.05) |
18.52 |
0.250(>0.05) |
|
>5 cm |
22.23 |
22.96 |
|||
|
Tumor location |
Right side |
21.14 |
0.478(>0.05) |
22.00 |
0.212(>0.05) |
|
Left side |
18.53 |
17.41 |
|||
There are no significant association between Stool and Serum MiR-21 expression level and clinical characteristics (age, genus, tumor type, tumor size and tumor location) of colorectal patients (P value >0.05).
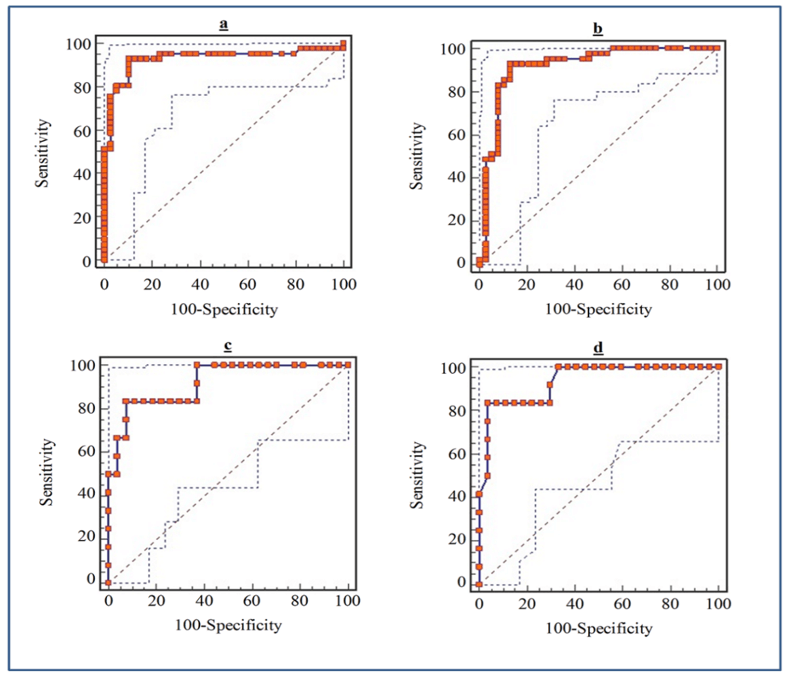
Figure 3: ROC curve of serum and stool miR-135b Expression level for detection of CRC patients. a) Serum miR-135b expression level for detection of CRC patients, area under the ROC curve (AUC): 0.929, Youden index: 0.8243, sensitivity: 92.7%, specificity: 89.7%. b) Stool miR-135b expression level for detection of CRC patients, AUC: 0.919, Youden index: 0.7986, Sensitivity: 92.7%, Specificity: 87.2% c) Serum miR-135b expression level for detection of TNM stages in CRC patients, AUC: 0.920, Youden index: 0.7593, sensitivity: 83.3%, specificity: 92.6%. d) Stool miR-135b expression level for detection of TNM stages in CRC patients, AUC: 0.935, Youden index: 0.7963, sensitivity: 83%, specificity: 92.5%.
II MiR-135b as a Noninvasive Biomarker for Diagnosis of Colorectal Cancer
To evaluate the diagnostic potential of miR-135b as a noninvasive biomarker, the ROC curves were studied (Figure 3). ROC curve analysis exhibited that serum miR-135b levels can be used as perfect diagnostic biomarker for CRC detection, with a sensitivity: 92.7%, specificity: 89.7%, AUC (area under the ROC curve) 0.929. Moreover, ROC analyses revealed that stool miR-135b levels discriminated CRC patients from control subjects with a sensitivity: 92.7%, specificity: 89.7%, AUC 0.984, with no significant statistic differences with serum miR-135b (P=0.72). The ROC analyses also, showed that serum miR-135b levels were robustious in detecting TNM stage III-IV from I-II with a sensitivity: 83.3% and specificity: 92.6%, AUC: 0.920. Furthermore, in patients with CRC, stool miR-135b levels, as well as serum miR-135b (P=0.7), were diagnosed TNM stage III-IV from I-II with a sensitivity: 83%, specificity: 92.5%, AUC: 0.935.
Discussion
In recent years, in order to achieve a safe and non-invasive method for the diagnosis of cancer, researchers have tended to consider micro-RNA profiles that are overexpressed in various cancers [11, 19, 28, 29]. Since the recognition of miRNAs in 1993, they have emerged as a favorable new class of biomarkers for cancer diagnosis [19, 31]. However, only a small number of miRNA are detectable in serum and stool, to the same extent that they are in the cancer cells [18, 32]. Hence, more studies should be done to specify how much the miRNAs are released into the bloodstream. Also, many of the cells in the colon cancer, in addition to the blood flow, drop their content into the lumen and sometimes the cells excreted in the stool which makes miRNAs released into the stool [14, 33, 34].
In this study, it was found that miR-135b in serum and stool of patients with colorectal cancer is upregulated, which confirms previous studies [35-39]. Statistical analysis of our study showed that the expression level of the miR-135b in serum and stool did not differ significantly, compared to the results of previous studies, which showed that miRNA expression is increased in tumor tissues [4, 34]. It can be said that the expression level of the miR-135b in serum, stool and tumor tissues of CRC patients is a correspondence of its expression in cancer cells. The other results of our study which examined the relationship between the expression of the miR-135b and some clinical and demographic characteristics showed that overexpression of the miR-135b in serum and stool of CRC patients was not significantly associated with age, gender, tumor size, tumor type and tumor location. Previous studies have shown that high levels of this miRNA had a positive significant relationship with the TNM stages [4, 34].
This study also revealed that miR-135b amount increased from the stage I to the IV, in the serum of CRC patients but in the stool, the expression level in stages III, IV had no significant difference. On the other hand, evaluating of ROC curve indicated that miR-135b expression in serum and stool can be used as an efficient diagnostic test to identify patients with colorectal cancer and determine the TNM stages of CRC patients with high sensitivity and specificity. The study of Wu.CW et al. (2014) showed that the sensitivity of fecal miR-135b for detection of colorectal cancer was 78% and the specificity was 68%, whereas in our study higher levels of sensitivity and specificity were acquired both in serum and stool. The comparison of ROC curves, using Medical statistical software (2009) indicated that the sensitivity and specificity of the miR-135b in serum and stool were not significantly different for CRC diagnosis and TNM stages determining.
Overall, the results of this study suggest that miR-135b in serum and stool have diagnostic value. Nonetheless, for efficiency analysis, the -ΔΔct equation was used, that is relative and does not indicate, the exact amount of miRNAs, and to get the correct results, it would need data normalization [40, 41]. Determination of appropriate internal control is the most important approach to normalizing the real-time PCR data [41]. Mostly, in previous studies, miR-16 and RNU6B were used as internal control, but until then there was no exact recommended miRNA as reference gene [42]. For this reason, we evaluate both miRNAs and it was found that miR-16 due to the greater stability was more suitable as an internal control. In summary, the results of the current study indicate that Serum and stool miR-135b expression levels seem to be a potential diagnostic biomarker for CRC patients. However, further well-designed studies with larger sample sizes are needed to well approve the role of miR-135b in CRC noninvasive diagnosis.
Acknowledgements
We appreciate the members of the Molecular medicine and Genetics department for their suggestions and assistance. This Study was supported by Hamadan University of Medical Sciences, Hamadan, Iran.
Conflicts of Interest
None.
Article Info
Article Type
Research ArticlePublication history
Received: Mon 06, Jan 2020Accepted: Mon 03, Feb 2020
Published: Fri 07, Feb 2020
Copyright
© 2023 Massoud Saidijam. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.CEI.2020.01.04
Author Info
Massoud Saidijam Morovat Taherikalani Nooshin Shabab Reza Ghanbari Saeid Afshar Saiyad Bastaminejad Salar Bakhtiyari
Corresponding Author
Massoud SaidijamDepartment of Molecular Medicine, School of Medicine, Hamadan University of Medical Sciences, Hamadan, Iran
Figures & Tables
Table 1: Patient’s clinical characteristics.
|
Healthy Controls |
Patients |
|
|
|
18 22 |
16 24 |
≤50 |
Age |
|
>50 |
|||
|
21 |
21 |
Male |
Genus |
|
19 |
19 |
Female |
|
|
|
11 |
I |
TNM staging |
|
|
16 |
II |
|
|
|
6 |
III |
|
|
|
7 |
IV |
|
|
|
32 |
Right side Colon |
Tumor location |
|
|
18 |
Left side colon |
These Patient Demographics information were extracted from patients Files in Shariati hospital.
Table 2: Inclusion and Exclusion Criteria to undergoing the study.
|
|
Patient |
Healthy control |
|
Inclusion Criteria |
A person that Was confirmed as malignant or precancerous by colonoscopy and histopathologic examination and was agreed to undergo the study. |
|
|
Exclusion Criteria |
A person who:
|
A person who:
|
Patients and healthy control group based on these criteria Were enrolled and the most of case histories were extracted from patient records.
Table 3: MiR-21 expression level in serum and stool, based on patients’ clinical characteristics.
|
|
|
MiR-21 in serum |
MiR-21 in stool |
||
|
Average rank |
P value |
Average rank |
P value |
||
|
Age |
≤50 |
20.37 |
0.688(>0.05) |
17.25 |
0.151(>0.05) |
|
>50 |
19.90 |
22.67 |
|||
|
Genus |
Male |
19.81 |
0.694(>0.05) |
20.52 |
0.989(>0.05) |
|
Female |
21.26 |
20.47 |
|||
|
Tumor type |
Colon |
21.09 |
0.52(>0.05) |
20.33 |
0.852(>0.05) |
|
Rectum |
18.12 |
20.19 |
|||
|
Tumor size |
≤5cm |
18.88 |
0.38 (>0.05) |
18.52 |
0.250(>0.05) |
|
>5 cm |
22.23 |
22.96 |
|||
|
Tumor location |
Right side |
21.14 |
0.478(>0.05) |
22.00 |
0.212(>0.05) |
|
Left side |
18.53 |
17.41 |
|||
There are no significant association between Stool and Serum MiR-21 expression level and clinical characteristics (age, genus, tumor type, tumor size and tumor location) of colorectal patients (P value >0.05).
References
- Siegel R, Desantis C, Jemal A (2014) Colorectal cancer statistics, 2014. CA Cancer J Clin 64: 104-17. [Crossref]
- Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65: 5-29. [Crossref]
- Wang S, Xiang J, Li Z, Lu S, Hu J et al. (2015) A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer 136: 152-161. [Crossref]
- Fearon ER (2011) Molecular genetics of colorectal cancer. Annu Rev Pathol 6: 479-507. [Crossref]
- Coppedè F, Lopomo A, Spisni R, Migliore L (2014) Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World J Gastroenterol 20: 943-956. [Crossref]
- Weng W, Feng J, Qin H, Ma Y, Goel A (2015) An update on miRNAs as biological and clinical determinants in colorectal cancer: a bench-to-bedside approach. Future Oncol 11: 1791-1808. [Crossref]
- Kraus S, Nabiochtchikov I, Shapira S, Arber N (2014) Recent advances in personalized colorectal cancer research. Cancer Lett 347: 15-21. [Crossref]
- Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K et al. (2013) Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst 105: 849-859. [Crossref]
- Hayes J, Peruzzi PP, Lawler S (2014) MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 20: 460-469. [Crossref]
- Kim M, Kasinski AL, Slack FJ (2011) MicroRNA therapeutics in preclinical cancer models. Lancet Oncol 12: 319-321. [Crossref]
- Yang X, Zhong J, Ji Y, Li J, Jian Y et al. (2015) The expression and clinical significance of microRNAs in colorectal cancer detecting. Tumour Biol 36: 2675-2684. [Crossref]
- Ellermeier C, Vang S, Cleveland K, Durand W, Resnick MB et al. (2014) Prognostic microRNA expression signature from examination of colorectal primary and metastatic tumors. Anticancer Res 34: 3957-3967. [Crossref]
- Mazeh H, Mizrahi I, Ilyayev N, Halle D, Brucher B et al. (2013) The Diagnostic and Prognostic Role of microRNA in Colorectal Cancer - a Comprehensive review. J Cancer 4: 281-295. [Crossref]
- Montano M (2011) MicroRNAs: miRRORS of health and disease. Transl Res 157: 157-162. [Crossref]
- Bartels CL, Tsongalis GJ (2009) MicroRNAs: novel biomarkers for human cancer. Clin Chem 55: 623-631. [Crossref]
- Liu R, Chen X, Du Y, Yao W, Shen L et al. (2012) Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem 58: 610-618. [Crossref]
- Luo X, Burwinkel B, Tao S, Brenner H (2011) MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev 20: 1272-1286. [Crossref]
- Li X, Zhang G, Luo F, Ruan J, Huang D et al. (2012) Identification of aberrantly expressed miRNAs in rectal cancer. Oncol Rep 28: 77-84. [Crossref]
- Jansson MD, Lund AH (2012) MicroRNA and cancer. Mol Oncol 6: 590-610. [Crossref]
- Afshar S, Najafi R, Sedighi Pashaki A, Sharifi M, Nikzad S et al. (2018) MiR-185 enhances radiosensitivity of colorectal cancer cells by targeting IGF1R and IGF2. Biomed Pharmacother 106: 763-769. [Crossref]
- Brunet Vega A, Pericay C, Moya I, Ferrer A, Dotor E et al. (2013) microRNA expression profile in stage III colorectal cancer: circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep 30: 320-326. [Crossref]
- Kanaan Z, Roberts H, Eichenberger MR, Billeter A, Ocheretner G et al. (2013) A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg 258: 400-408. [Crossref]
- Koga Y, Yasunaga M, Takahashi A, Kuroda J, Moriya Y et al. (2010) MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev Res (Phila) 3: 1435-1442. [Crossref]
- Sarver AL, French AJ, Borralho PM, Thayanithy V, Oberg AL et al. (2009) Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer 9: 401. [Crossref]
- Saito T, Saetrom P (2010) MicroRNAs--targeting and target prediction. New Biotechnol 27: 243-249. [Crossref]
- Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA et al. (2008) Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res 68: 5795-5802. [Crossref]
- Becker C, Hammerle Fickinger A, Riedmaier I, Pfaffl MW (2010) mRNA and microRNA quality control for RT-qPCR analysis. Methods 50: 237-243. [Crossref]
- Kroh EM, Parkin RK, Mitchell PS, Tewari M (2010) Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 50: 298-301. [Crossref]
- Jacobsen N, Andreasen D, Mouritzen P (2011) Profiling microRNAs by real-time PCR. Methods Mol Biol 732: 39-54. [Crossref]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402-408. [Crossref]
- Yong FL, Law CW, Wang CW (2013) Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer 13: 280. [Crossref]
- Zen K, Zhang CY (2012) Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev 32: 326-348. [Crossref]
- Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME (2004) Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med 351: 2704-2714. [Crossref]
- Wu CW, Ng SC, Dong Y, Tian L, Ng SS et al. (2014) Identification of microRNA-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin Cancer Res 20: 2994-3002. [Crossref]
- Zanutto S, Pizzamiglio S, Ghilotti M, Bertan C, Ravagnani F et al. (2014) Circulating miR-378 in plasma: a reliable, haemolysis-independent biomarker for colorectal cancer. Br J Cancer 110: 1001-1007. [Crossref]
- Ahlquist DA, Kisiel JB (2015) Stool DNA for Colorectal Cancer Screening: From Concepts to Quality Care. In: Shaukat A, Allen JI, editors. Colorectal Cancer Screening: Quality and Benchmarks. Springer New York 97-112.
- Brase JC, Wuttig D, Kuner R, Sultmann H (2010) Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer 9: 306. [Crossref]
- Schetter AJ, Okayama H, Harris CC (2012) The role of microRNAs in colorectal cancer. Cancer J 18: 244-252. [Crossref]
- Iorio MV, Croce CM (2012) MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4: 143-159. [Crossref]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nature Protoc 3: 1101-1108. [Crossref]
- Mar JC, Kimura Y, Schroder K, Irvine KM, Hayashizaki Y et al. (2009) Data-driven normalization strategies for high-throughput quantitative RT-PCR. BMC Bioinformatics 10: 110. [Crossref]
- Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ et al. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev 19: 1766-1774. [Crossref]
